Using `WeightIt` R package for causal inference analyses
I recently discovered WeightIt
R package and was very happy with its functionality and performance. I “delegated” my code computing IPTW to WeightIt and it was faster while producing the same results, as expected. In this post I will use publicly available data to demonstrate the great functionality of WeightIt and the scope of challenges it may help addressing.
Open data
temp <- tempfile()
temp2 <- tempfile()
download.file("https://cdn1.sph.harvard.edu/wp-content/uploads/sites/1268/2017/01/nhefs_excel.zip", temp)
unzip(zipfile = temp, exdir = temp2)
data <- readxl::read_xls(file.path(temp2, "NHEFS.xls"))
unlink(c(temp, temp2))
data
## # A tibble: 1,629 × 64
## seqn qsmk death yrdth modth dadth sbp dbp sex age race income
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 233 0 0 NA NA NA 175 96 0 42 1 19
## 2 235 0 0 NA NA NA 123 80 0 36 0 18
## 3 244 0 0 NA NA NA 115 75 1 56 1 15
## 4 245 0 1 85 2 14 148 78 0 68 1 15
## 5 252 0 0 NA NA NA 118 77 0 40 0 18
## 6 257 0 0 NA NA NA 141 83 1 43 1 11
## 7 262 0 0 NA NA NA 132 69 1 56 0 19
## 8 266 0 0 NA NA NA 100 53 1 29 0 22
## 9 419 0 1 84 10 13 163 79 0 51 0 18
## 10 420 0 1 86 10 17 184 106 0 43 0 16
## # … with 1,619 more rows, and 52 more variables: marital <dbl>, school <dbl>,
## # education <dbl>, ht <dbl>, wt71 <dbl>, wt82 <dbl>, wt82_71 <dbl>,
## # birthplace <dbl>, smokeintensity <dbl>, smkintensity82_71 <dbl>,
## # smokeyrs <dbl>, asthma <dbl>, bronch <dbl>, tb <dbl>, hf <dbl>, hbp <dbl>,
## # pepticulcer <dbl>, colitis <dbl>, hepatitis <dbl>, chroniccough <dbl>,
## # hayfever <dbl>, diabetes <dbl>, polio <dbl>, tumor <dbl>,
## # nervousbreak <dbl>, alcoholpy <dbl>, alcoholfreq <dbl>, …
For this section, I will partly re-use the code by Joy Shi and Sean McGrath available here.
data %<>%
mutate(
# define censoring
cens = if_else(is.na(wt82_71),1, 0),
# categorize the school variable (as in R code by Joy Shi and Sean McGrath)
education = cut(school, breaks = c(0, 8, 11, 12, 15, 20),
include.lowest = TRUE,
labels = c('1. 8th Grage or Less',
'2. HS Dropout',
'3. HS',
'4. College Dropout',
'5. College or More')),
# establish active as a factor variable
active = factor(active),
# establish exercise as a factor variable
exercise = factor(exercise),
# create a treatment label variable
qsmklabel = if_else(qsmk == 1,
'Quit Smoking 1971-1982',
'Did Not Quit Smoking 1971-1982'),
# survtime variable
survtime = if_else(death == 0, 120, (yrdth-83)*12 + modth) # yrdth ranges from 83 to 92
) %>%
# ignore those with missing values on some variables
filter(!is.na(education))
Descriptive table with balancing characteristics
table <- bal.tab(death ~ qsmk + sex + race + age + education + smokeintensity + smkintensity82_71 + smokeyrs + exercise + active + wt71, data = data, estimand = "ATE", thresholds = c(m = 0.1))
table
## Balance Measures
## Type Diff.Un M.Threshold.Un
## qsmk Binary 0.0721 Balanced, <0.1
## sex Binary -0.1525 Not Balanced, >0.1
## race Binary 0.0275 Balanced, <0.1
## age Contin. 1.3419 Not Balanced, >0.1
## education_1. 8th Grage or Less Binary 0.2356 Not Balanced, >0.1
## education_2. HS Dropout Binary 0.0136 Balanced, <0.1
## education_3. HS Binary -0.1705 Not Balanced, >0.1
## education_4. College Dropout Binary -0.0297 Balanced, <0.1
## education_5. College or More Binary -0.0490 Balanced, <0.1
## smokeintensity Contin. 0.0195 Balanced, <0.1
## smkintensity82_71 Contin. -0.2598 Not Balanced, >0.1
## smokeyrs Contin. 1.2315 Not Balanced, >0.1
## exercise_0 Binary -0.0464 Balanced, <0.1
## exercise_1 Binary -0.0788 Balanced, <0.1
## exercise_2 Binary 0.1252 Not Balanced, >0.1
## active_0 Binary -0.0950 Balanced, <0.1
## active_1 Binary 0.0466 Balanced, <0.1
## active_2 Binary 0.0484 Balanced, <0.1
## wt71 Contin. 0.1181 Not Balanced, >0.1
##
## Balance tally for mean differences
## count
## Balanced, <0.1 11
## Not Balanced, >0.1 8
##
## Variable with the greatest mean difference
## Variable Diff.Un M.Threshold.Un
## age 1.3419 Not Balanced, >0.1
##
## Sample sizes
## Control Treated
## All 1311 318
Several important covariables are unbalanced with standardized mean difference > 10% (SMD > 0.1), e.g., sex, age, education, previous weight wt71, previous smoking intensity and duration, and psychical activity level. Not adressing confounding by these factors, we cannot estimate the true average causal effect (ATE) of smoking cessation qsmk on mortality
Computing inverse probability of treatment weights for the point treatment
iptw <- weightit(qsmk ~ sex + race + age + I(age*age) + as.factor(education) + smokeintensity + I(smokeintensity*smokeintensity) + smokeyrs + I(smokeyrs*smokeyrs) + as.factor(exercise) + as.factor(active) + wt71 + I(wt71*wt71), data = data, estimand = "ATE", method = "ps")
iptw
## A weightit object
## - method: "ps" (propensity score weighting)
## - number of obs.: 1629
## - sampling weights: none
## - treatment: 2-category
## - estimand: ATE
## - covariates: sex, race, age, I(age * age), as.factor(education), smokeintensity, I(smokeintensity * smokeintensity), smokeyrs, I(smokeyrs * smokeyrs), as.factor(exercise), as.factor(active), wt71, I(wt71 * wt71)
summary(iptw)
## Summary of weights
##
## - Weight ranges:
##
## Min Max
## treated 1.2607 |---------------------------| 16.0062
## control 1.0559 |--| 2.9229
##
## - Units with 5 most extreme weights by group:
##
## 1551 1503 1244 1376 170
## treated 10.1178 11.0187 12.3286 15.82 16.0062
## 722 633 774 1229 1088
## control 2.5007 2.5423 2.6693 2.8155 2.9229
##
## - Weight statistics:
##
## Coef of Var MAD Entropy # Zeros
## treated 0.475 0.334 0.095 0
## control 0.171 0.122 0.013 0
##
## - Effective Sample Sizes:
##
## Control Treated
## Unweighted 1201. 428.
## Weighted 1166.82 349.46
There are no extreme weights; the exposed vs unexposed counts in the raw and weighted populations are presented. Now we can investigate how much we improved the exchangeability between exposed and unexposed by re-weighting the original population using computed IPTW.
table_iptw <- bal.tab(iptw, stats = c("m"), thresholds = c(m = 0.01))
table_iptw
## Call
## weightit(formula = qsmk ~ sex + race + age + I(age * age) + as.factor(education) +
## smokeintensity + I(smokeintensity * smokeintensity) + smokeyrs +
## I(smokeyrs * smokeyrs) + as.factor(exercise) + as.factor(active) +
## wt71 + I(wt71 * wt71), data = data, method = "ps", estimand = "ATE")
##
## Balance Measures
## Type Diff.Adj M.Threshold
## prop.score Distance 0.0081 Balanced, <0.01
## sex Binary -0.0010 Balanced, <0.01
## race Binary 0.0025 Balanced, <0.01
## age Contin. 0.0022 Balanced, <0.01
## I(age * age) Contin. 0.0023 Balanced, <0.01
## as.factor(education)_1. 8th Grage or Less Binary -0.0093 Balanced, <0.01
## as.factor(education)_2. HS Dropout Binary 0.0013 Balanced, <0.01
## as.factor(education)_3. HS Binary 0.0011 Balanced, <0.01
## as.factor(education)_4. College Dropout Binary 0.0059 Balanced, <0.01
## as.factor(education)_5. College or More Binary 0.0009 Balanced, <0.01
## smokeintensity Contin. -0.0226 Not Balanced, >0.01
## I(smokeintensity * smokeintensity) Contin. -0.0254 Not Balanced, >0.01
## smokeyrs Contin. -0.0040 Balanced, <0.01
## I(smokeyrs * smokeyrs) Contin. -0.0013 Balanced, <0.01
## as.factor(exercise)_0 Binary -0.0038 Balanced, <0.01
## as.factor(exercise)_1 Binary 0.0188 Not Balanced, >0.01
## as.factor(exercise)_2 Binary -0.0149 Not Balanced, >0.01
## as.factor(active)_0 Binary -0.0064 Balanced, <0.01
## as.factor(active)_1 Binary 0.0122 Not Balanced, >0.01
## as.factor(active)_2 Binary -0.0058 Balanced, <0.01
## wt71 Contin. 0.0019 Balanced, <0.01
## I(wt71 * wt71) Contin. 0.0047 Balanced, <0.01
##
## Balance tally for mean differences
## count
## Balanced, <0.01 17
## Not Balanced, >0.01 5
##
## Variable with the greatest mean difference
## Variable Diff.Adj M.Threshold
## I(smokeintensity * smokeintensity) -0.0254 Not Balanced, >0.01
##
## Effective sample sizes
## Control Treated
## Unadjusted 1201. 428.
## Adjusted 1166.82 349.46
An important covariable smokeintensity remained unbalanced; however, there is a clear exchangeability improvement (less confounding) in the re-weighted population vs original one (wrt sex, age, physical activity variables and previous weight wt71).
We can run a weighted Cox proportional hazards regression model using computed weights.
# crude Cox PH model
cox_model_crude <- coxph(Surv(survtime, death == 1) ~ qsmk, data = data)
cox_fit_crude <- cox_model_crude %>% broom::tidy(., conf.int = T, exponentiate = T)
# IPTW Cox PH model
cox_model_iptw <- coxph(Surv(survtime, death == 1) ~ qsmk, data = data, weights = iptw$weights)
cox_fit_iptw <- cox_model_iptw %>% broom::tidy(., conf.int = T, exponentiate = T)
# HR
cox_fit_crude %>% bind_rows(cox_fit_iptw)
## # A tibble: 2 × 8
## term estimate std.error statistic p.value conf.low conf.high robust.se
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 qsmk 1.39 0.120 2.77 0.00565 1.10 1.76 NA
## 2 qsmk 1.00 0.0795 0.00883 0.993 0.770 1.30 0.134
Next, I compute 95% CIs using bootsrap percentiles.
library(tidymodels)
## Registered S3 method overwritten by 'tune':
## method from
## required_pkgs.model_spec parsnip
## ── Attaching packages ────────────────────────────────────── tidymodels 0.1.4 ──
## ✓ broom 0.7.10 ✓ rsample 0.1.1
## ✓ dials 0.0.10 ✓ tune 0.1.6
## ✓ infer 1.0.0 ✓ workflows 0.2.4
## ✓ modeldata 0.1.1 ✓ workflowsets 0.1.0
## ✓ parsnip 0.1.7 ✓ yardstick 0.0.9
## ✓ recipes 0.1.17
## ── Conflicts ───────────────────────────────────────── tidymodels_conflicts() ──
## x scales::discard() masks purrr::discard()
## x magrittr::extract() masks tidyr::extract()
## x dplyr::filter() masks stats::filter()
## x recipes::fixed() masks stringr::fixed()
## x dplyr::lag() masks stats::lag()
## x rsample::permutations() masks gtools::permutations()
## x MASS::select() masks dplyr::select()
## x magrittr::set_names() masks purrr::set_names()
## x yardstick::spec() masks readr::spec()
## x recipes::step() masks stats::step()
## • Use tidymodels_prefer() to resolve common conflicts.
times <- 1e3 # 1K samples
set.seed(876434567)
boots <- bootstraps(data, times = times, apparent = FALSE)
# function to plug-in the sampled data sets
cox_model <- function(data, iptw) {
coxph(Surv(survtime, death == 1) ~ qsmk, data = data, weights = iptw$weights)
}
boot_models <- boots %>%
mutate(
# explicitly transform sampled datasets/splits to tibbles
splits = map(splits, ~as_tibble(.x)),
# compute IPTW in each split/sampled data set
iptw = map(splits, ~weightit(qsmk ~ sex + race + age + I(age*age) + as.factor(education) + smokeintensity + I(smokeintensity*smokeintensity) + smokeyrs + I(smokeyrs*smokeyrs) + as.factor(exercise) + as.factor(active) + wt71 + I(wt71*wt71), data = .x, estimand = "ATE", method = "ps")),
# using each data sample and corresponding IPTWs, run weighted Cox PH model
model = map2(.x = splits, .y = iptw, ~cox_model(data = .x, iptw = .y)),
coef_info = map(model, ~broom::tidy(.x, exponentiate = T)))
# combine coefficients from all sampled data sets in one tibble
boot_coefs <- boot_models %>%
unnest(coef_info)
# compute 95% CI using all sampled data sets results
percentile_intervals <- boot_coefs %>%
summarise_at(.vars = vars(estimate),
.funs = list(Q2.5 = ~ quantile(., probs = 0.025), # lower bound of 95% CI
Q50 = ~ quantile(., probs = 0.50), # point estimate
Q97.5 = ~ quantile(., probs = 0.975))) # upper bound of 95% CI
percentile_intervals
## # A tibble: 1 × 3
## Q2.5 Q50 Q97.5
## <dbl> <dbl> <dbl>
## 1 0.802 0.998 1.24
Overall, we found no effect of smoking cessation on mortality at the end of data at t=120 months.
Simulated data
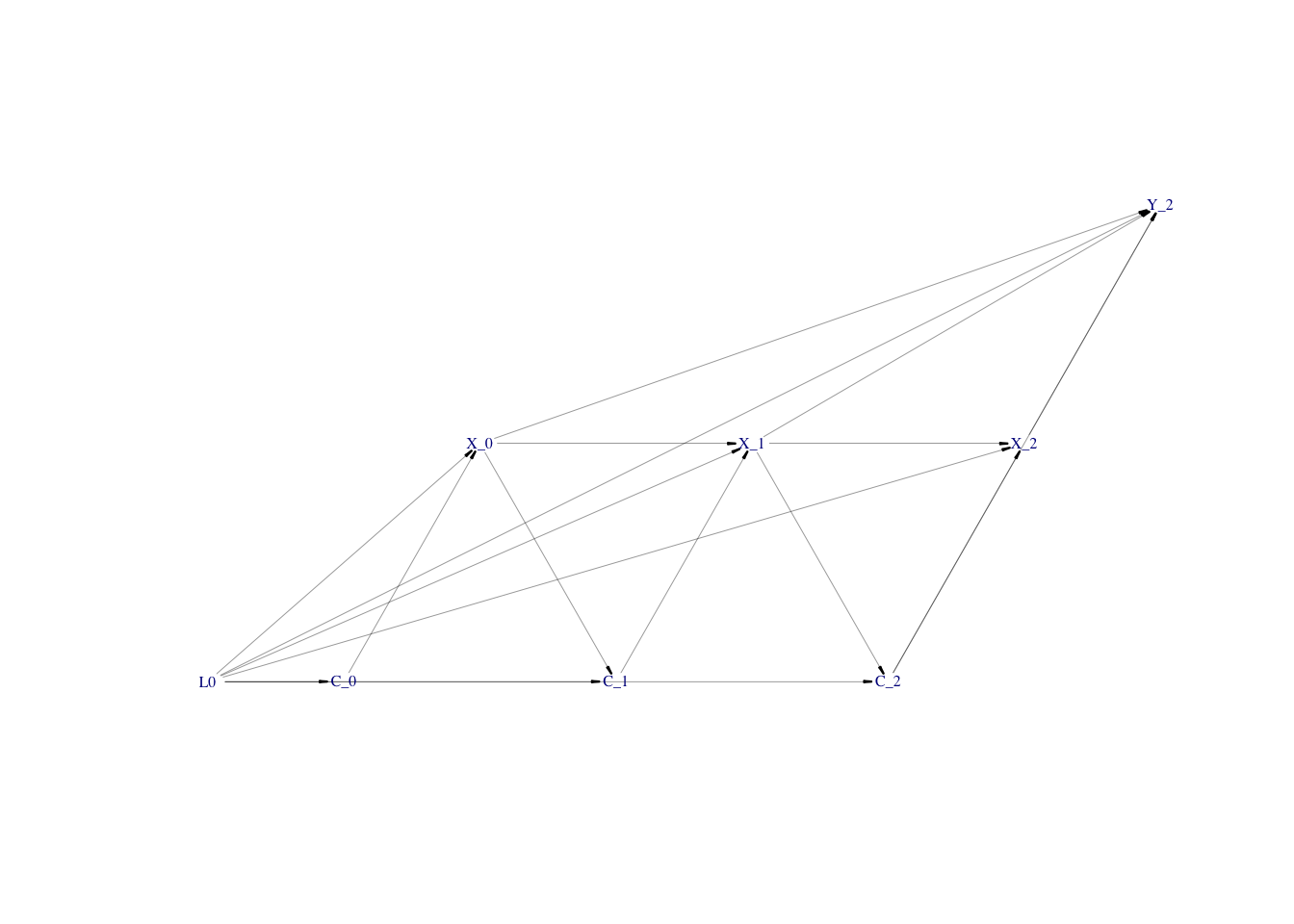
First, I will use the example of the simulated time-varying confounding and exposure using simcausal
package vignette. I reduce the number of time points to 3 (see the final directed acyclic graph presenting the simulated data), simulate one cumulative outcome at the end of data at t=2 and add effects of previous exposures X on the cumulative outcome at t=2 (see DAG).
library(simcausal)
D <- DAG.empty() +
node("L0", distr = "rcat.b1", probs = c(0.5, 0.25, 0.25)) +
node("C", t = 0, distr = "rnorm", mean = 3 + (L0 > 1) * 3 + (L0 > 2) * 3) +
node("X", t = 0, distr = "rbern", prob = plogis(0.2 + 0.3 * L0 + 0.5 * C[t])) +
node("C", t = 1:2, distr = "rnorm", mean = -X[t-1]*10 + 5 + (L0 > 1) * 3 + (L0 > 2) * 2) +
node("X", t = 1:2, distr = "rbern", prob = plogis(0.1 + 0.1 * L0 + 0.4 * C[t] + 2 * X[t - 1])) +
node("Y", t = 2, distr = "rbern", prob = plogis(0.1 + 1.2 * X[t] + 1.3 * X[t-1] + 1.1 * X[t-2] + 0.1 * L0 + 0.2 * C[t]), EFU = TRUE)
D <- set.DAG(D)
## ...automatically assigning order attribute to some nodes...
## node L0, order:1
## node C_0, order:2
## node X_0, order:3
## node C_1, order:4
## node X_1, order:5
## node C_2, order:6
## node X_2, order:7
## node Y_2, order:8
plotDAG(D)
## using the following vertex attributes:
## NAdarkbluenone70.50
## using the following edge attributes:
## black0.210.30.2
dat.long <- sim(D, n = 3e3)
## simulating observed dataset from the DAG object
# NAs are not allowed for exposure nodes; I want to code L0 explicitly as a factor with 3 levels and not as continuous variable
dat.long %<>%
filter(!is.na(X_0)) %>%
filter(!is.na(X_1)) %>%
filter(!is.na(X_2)) %>%
mutate(
L0 = as.factor(L0)
)
Using WeightIt, I will compute the total effect of exposure X at t=0, t=1 and t=2 on the outcome Y while adjusting for the time-varying confounder C. But first, I check the covariables balance at every time-point in the time-varying simulated data set.
balance_tab <- bal.tab(list(X_0 ~ L0 + C_0,
X_1 ~ X_0 + L0 + C_1,
X_2 ~ X_0 + X_1 + L0 + C_1 + C_2),
data = dat.long, stats = c("m"), thresholds = c(m = 0.1),
which.time = .all)
balance_tab
## Balance by Time Point
##
## - - - Time: 1 - - -
## Balance Measures
## Type Diff.Un M.Threshold.Un
## L0_1 Binary -0.4477 Not Balanced, >0.1
## L0_2 Binary 0.1983 Not Balanced, >0.1
## L0_3 Binary 0.2494 Not Balanced, >0.1
## C_0 Contin. 1.1548 Not Balanced, >0.1
##
## Balance tally for mean differences
## count
## Balanced, <0.1 0
## Not Balanced, >0.1 4
##
## Variable with the greatest mean difference
## Variable Diff.Un M.Threshold.Un
## C_0 1.1548 Not Balanced, >0.1
##
## Sample sizes
## Control Treated
## All 222 2778
##
## - - - Time: 2 - - -
## Balance Measures
## Type Diff.Un M.Threshold.Un
## X_0 Binary -0.0654 Balanced, <0.1
## L0_1 Binary -0.3346 Not Balanced, >0.1
## L0_2 Binary 0.0939 Balanced, <0.1
## L0_3 Binary 0.2408 Not Balanced, >0.1
## C_1 Contin. 0.9073 Not Balanced, >0.1
##
## Balance tally for mean differences
## count
## Balanced, <0.1 2
## Not Balanced, >0.1 3
##
## Variable with the greatest mean difference
## Variable Diff.Un M.Threshold.Un
## C_1 0.9073 Not Balanced, >0.1
##
## Sample sizes
## Control Treated
## All 826 2174
##
## - - - Time: 3 - - -
## Balance Measures
## Type Diff.Un M.Threshold.Un
## X_0 Binary 0.0722 Balanced, <0.1
## X_1 Binary -0.2291 Not Balanced, >0.1
## L0_1 Binary -0.2453 Not Balanced, >0.1
## L0_2 Binary 0.0482 Balanced, <0.1
## L0_3 Binary 0.1971 Not Balanced, >0.1
## C_1 Contin. 0.0915 Balanced, <0.1
## C_2 Contin. 0.9404 Not Balanced, >0.1
##
## Balance tally for mean differences
## count
## Balanced, <0.1 3
## Not Balanced, >0.1 4
##
## Variable with the greatest mean difference
## Variable Diff.Un M.Threshold.Un
## C_2 0.9404 Not Balanced, >0.1
##
## Sample sizes
## Control Treated
## All 634 2366
## - - - - - - - - - - -
Evidently, the farther observations are from the baseline t=0, the more imbalance there is.
Next, I will construct PS models for every time point in the data set to compute IPTWs.
iptw_models <- weightitMSM(list(X_0 ~ L0 + C_0,
X_1 ~ X_0 + L0 + C_1,
X_2 ~ X_0 + X_1 + L0 + C_1 + C_2),
data = dat.long, method = "ps", stabilize = TRUE)
iptw_models
## A weightitMSM object
## - method: "ps" (propensity score weighting)
## - number of obs.: 3000
## - sampling weights: none
## - number of time points: 3 (X_0, X_1, X_2)
## - treatment:
## + time 1: 2-category
## + time 2: 2-category
## + time 3: 2-category
## - covariates:
## + baseline: L0, C_0
## + after time 1: X_0, L0, C_1
## + after time 2: X_0, X_1, L0, C_1, C_2
## - stabilized; stabilization factors:
## + baseline: (none)
## + after time 1: X_0
## + after time 2: X_0, X_1, X_0:X_1
summary(iptw_models)
## Summary of weights
##
## Time 1
## Summary of weights
##
## - Weight ranges:
##
## Min Max
## treated 0.2488 |--------------------| 9.0953
## control 0.1173 |---------------------------| 11.3726
##
## - Units with 5 most extreme weights by group:
##
## 1492 389 1916 392 426
## treated 5.1977 5.4518 5.4809 6.0153 9.0953
## 1140 71 130 149 950
## control 6.5338 6.9171 8.839 9.6473 11.3726
##
## - Weight statistics:
##
## Coef of Var MAD Entropy # Zeros
## treated 0.650 0.450 0.158 0
## control 1.605 0.773 0.600 0
##
## - Mean of Weights = 0.99
##
## - Effective Sample Sizes:
##
## Control Treated
## Unweighted 222. 2778.
## Weighted 62.27 1953.57
##
## Time 2
## Summary of weights
##
## - Weight ranges:
##
## Min Max
## treated 0.1173 |---------------------------| 11.3726
## control 0.1215 |---------------------| 9.0953
##
## - Units with 5 most extreme weights by group:
##
## 1140 71 130 149 950
## treated 6.5338 6.9171 8.839 9.6473 11.3726
## 1492 389 1916 392 426
## control 5.1977 5.4518 5.4809 6.0153 9.0953
##
## - Weight statistics:
##
## Coef of Var MAD Entropy # Zeros
## treated 0.711 0.462 0.173 0
## control 0.833 0.503 0.227 0
##
## - Mean of Weights = 0.99
##
## - Effective Sample Sizes:
##
## Control Treated
## Unweighted 826. 2174.
## Weighted 487.9 1443.92
##
## Time 3
## Summary of weights
##
## - Weight ranges:
##
## Min Max
## treated 0.1215 |-----------------------| 9.6473
## control 0.1173 |---------------------------| 11.3726
##
## - Units with 5 most extreme weights by group:
##
## 1140 71 130 426 149
## treated 6.5338 6.9171 8.839 9.0953 9.6473
## 2363 1666 65 1688 950
## control 3.5002 3.5116 3.523 5.2801 11.3726
##
## - Weight statistics:
##
## Coef of Var MAD Entropy # Zeros
## treated 0.749 0.493 0.193 0
## control 0.728 0.397 0.166 0
##
## - Mean of Weights = 0.99
##
## - Effective Sample Sizes:
##
## Control Treated
## Unweighted 634. 2366.
## Weighted 414.75 1515.35
No extreme weights were produced. Now I will check the covariables balance when the population is reweighted at each time point by the IPT weights computed above.
balance_tab_iptw <- bal.tab(iptw_models, stats = c("m"), thresholds = c(m = 0.1),
which.time = .all)
balance_tab_iptw
## Call
## weightitMSM(formula.list = list(X_0 ~ L0 + C_0, X_1 ~ X_0 + L0 +
## C_1, X_2 ~ X_0 + X_1 + L0 + C_1 + C_2), data = dat.long,
## method = "ps", stabilize = TRUE)
##
## Balance by Time Point
##
## - - - Time: 1 - - -
## Balance Measures
## Type Diff.Adj M.Threshold
## prop.score Distance 0.0837 Balanced, <0.1
## L0_1 Binary -0.0443 Balanced, <0.1
## L0_2 Binary -0.0261 Balanced, <0.1
## L0_3 Binary 0.0705 Balanced, <0.1
## C_0 Contin. 0.1405 Not Balanced, >0.1
##
## Balance tally for mean differences
## count
## Balanced, <0.1 4
## Not Balanced, >0.1 1
##
## Variable with the greatest mean difference
## Variable Diff.Adj M.Threshold
## C_0 0.1405 Not Balanced, >0.1
##
## Effective sample sizes
## Control Treated
## Unadjusted 222. 2778.
## Adjusted 62.27 1953.57
##
## - - - Time: 2 - - -
## Balance Measures
## Type Diff.Adj M.Threshold
## prop.score Distance 0.1068
## X_0 Binary -0.0707 Balanced, <0.1
## L0_1 Binary -0.0049 Balanced, <0.1
## L0_2 Binary 0.0043 Balanced, <0.1
## L0_3 Binary 0.0006 Balanced, <0.1
## C_1 Contin. 0.2591 Not Balanced, >0.1
##
## Balance tally for mean differences
## count
## Balanced, <0.1 4
## Not Balanced, >0.1 1
##
## Variable with the greatest mean difference
## Variable Diff.Adj M.Threshold
## C_1 0.2591 Not Balanced, >0.1
##
## Effective sample sizes
## Control Treated
## Unadjusted 826. 2174.
## Adjusted 487.9 1443.92
##
## - - - Time: 3 - - -
## Balance Measures
## Type Diff.Adj M.Threshold
## prop.score Distance 0.4357
## X_0 Binary 0.0452 Balanced, <0.1
## X_1 Binary -0.2512 Not Balanced, >0.1
## L0_1 Binary -0.0258 Balanced, <0.1
## L0_2 Binary -0.0169 Balanced, <0.1
## L0_3 Binary 0.0426 Balanced, <0.1
## C_1 Contin. -0.0709 Balanced, <0.1
## C_2 Contin. 0.6987 Not Balanced, >0.1
##
## Balance tally for mean differences
## count
## Balanced, <0.1 5
## Not Balanced, >0.1 2
##
## Variable with the greatest mean difference
## Variable Diff.Adj M.Threshold
## C_2 0.6987 Not Balanced, >0.1
##
## Effective sample sizes
## Control Treated
## Unadjusted 634. 2366.
## Adjusted 414.75 1515.35
## - - - - - - - - - - -
Although IPT weighting meaningfully improved covariables' balance, for confounder C at t=1, SMD indicated distribution imbalance for confounder C at t=2 and several other variables, which serves as the evidence of corrupted conditional exchangeability (residual confounding is present).
library(survey)
## Loading required package: grid
## Loading required package: Matrix
##
## Attaching package: 'Matrix'
## The following objects are masked from 'package:tidyr':
##
## expand, pack, unpack
##
## Attaching package: 'survey'
## The following object is masked from 'package:graphics':
##
## dotchart
res_svy <- svydesign(~1, weights = iptw_models$weights, data = dat.long)
res_svy_full <- svyglm(Y_2 ~ X_0 * X_1 * X_2, design = res_svy)
res_svy_full %>% broom::tidy(exponentiate = T, conf.int = T)
## # A tibble: 8 × 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 2.72 2.36e-13 4.24e+12 0 2.72 2.72
## 2 X_0 0.972 2.10e- 2 -1.38e+ 0 0.169 0.932 1.01
## 3 X_1 0.841 5.13e- 2 -3.37e+ 0 0.000749 0.761 0.930
## 4 X_2 0.954 3.69e- 2 -1.27e+ 0 0.204 0.888 1.03
## 5 X_0:X_1 1.06 5.79e- 2 1.01e+ 0 0.313 0.946 1.19
## 6 X_0:X_2 1.05 4.29e- 2 1.11e+ 0 0.266 0.964 1.14
## 7 X_1:X_2 1.14 6.94e- 2 1.82e+ 0 0.0681 0.991 1.30
## 8 X_0:X_1:X_2 0.985 7.49e- 2 -1.97e- 1 0.844 0.851 1.14
main_effects <- svyglm(Y_2 ~ X_0 + X_1 + X_2, design = res_svy)
main_effects %>% broom::tidy(exponentiate = T, conf.int = T)
## # A tibble: 4 × 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 2.29 0.0286 28.9 1.96e-162 2.16 2.42
## 2 X_0 1.05 0.0248 2.04 4.16e- 2 1.00 1.10
## 3 X_1 0.988 0.00807 -1.48 1.40e- 1 0.973 1.00
## 4 X_2 1.11 0.0156 6.39 1.88e- 10 1.07 1.14
cum_fit <- svyglm(Y_2 ~ I(X_0 + X_1 + X_2), design = res_svy)
cum_fit %>% broom::tidy(exponentiate = T, conf.int = T)
## # A tibble: 2 × 7
## term estimate std.error statistic p.value conf.low conf.high
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 2.32 0.0216 39.0 2.16e-269 2.22 2.42
## 2 I(X_0 + X_1 + X_2) 1.04 0.00816 5.19 2.21e- 7 1.03 1.06
On the relative scale, the cumulative effect of 3 doses of exposure X was associated with 4% increased risk of Y at the time t=2 (end of data). Had this analysis been based on non-simulated data, the detailed interpretation of this measure of effect would depend on the research question and domain expertise. Since this was an example using simulated data set with no true underlying clinical or otherwise practical and socially important research question, I get to claim that 4% increased risk does not warranty any concern.
Take home message
WeightIt
R package is great. It increased my quality of research life by sparing me the additional “by hand” coding. I am sure it is and will be useful for many research folks out there ✌️
Cheers! ✌️
References
- Hernán MA, Robins JM (2020). Causal Inference: What If. Boca Raton: Chapman & Hall/CRC
- R code by Joy Shi and Sean McGrath available here
WeightItR package: https://cran.r-project.org/web/packages/WeightIt/index.htmlsimcausalR package: https://github.com/osofr/simcausal- Austin, Peter C. 2011. “An Introduction to Propensity Score Methods for Reducing the Effects of Confounding in Observational Studies.” Multivariate Behavioral Research 46 (3): 399–424. https://doi.org/10.1080/00273171.2011.568786.
- Austin, Peter C., and Elizabeth A. Stuart. 2015. “Moving Towards Best Practice When Using Inverse Probability of Treatment Weighting (IPTW) Using the Propensity Score to Estimate Causal Treatment Effects in Observational Studies.” Statistics in Medicine 34 (28): 3661–79. https://doi.org/10.1002/sim.6607