In this post I will explore the use of medications, which are indicated to stop migraine episodes or to prevent them. I will use publicly available aggregated data on prescribed medication in Denmark between 1996 and 2019.
library(tidyverse)
library(magrittr)
library(colorfindr)
library(ggrepel)
Danish data
Getting the data
link_list_dk <- list(
"1996_atc_code_data.txt" = "https://medstat.dk/da/download/file/MTk5Nl9hdGNfY29kZV9kYXRhLnR4dA==",
"1997_atc_code_data.txt" = "https://medstat.dk/da/download/file/MTk5N19hdGNfY29kZV9kYXRhLnR4dA==",
"1998_atc_code_data.txt" = "https://medstat.dk/da/download/file/MTk5OF9hdGNfY29kZV9kYXRhLnR4dA==",
"1999_atc_code_data.txt" = "https://medstat.dk/da/download/file/MTk5OV9hdGNfY29kZV9kYXRhLnR4dA==",
"2000_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwMF9hdGNfY29kZV9kYXRhLnR4dA==",
"2001_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwMV9hdGNfY29kZV9kYXRhLnR4dA==",
"2002_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwMl9hdGNfY29kZV9kYXRhLnR4dA==",
"2003_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwM19hdGNfY29kZV9kYXRhLnR4dA==",
"2004_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwNF9hdGNfY29kZV9kYXRhLnR4dA==",
"2006_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwNl9hdGNfY29kZV9kYXRhLnR4dA==",
"2007_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwN19hdGNfY29kZV9kYXRhLnR4dA==",
"2008_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwOF9hdGNfY29kZV9kYXRhLnR4dA==",
"2009_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAwOV9hdGNfY29kZV9kYXRhLnR4dA==",
"2010_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxMF9hdGNfY29kZV9kYXRhLnR4dA==",
"2011_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxMV9hdGNfY29kZV9kYXRhLnR4dA==",
"2012_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxMl9hdGNfY29kZV9kYXRhLnR4dA==",
"2013_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxM19hdGNfY29kZV9kYXRhLnR4dA==",
"2014_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxNF9hdGNfY29kZV9kYXRhLnR4dA==",
"2015_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxNV9hdGNfY29kZV9kYXRhLnR4dA==",
"2016_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxNl9hdGNfY29kZV9kYXRhLnR4dA==",
"2017_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxN19hdGNfY29kZV9kYXRhLnR4dA==",
"2018_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxOF9hdGNfY29kZV9kYXRhLnR4dA==",
"2019_atc_code_data.txt" = "https://medstat.dk/da/download/file/MjAxOV9hdGNfY29kZV9kYXRhLnR4dA==",
"atc_code_text.txt" = "https://medstat.dk/da/download/file/YXRjX2NvZGVfdGV4dC50eHQ=",
"atc_groups.txt" = "https://medstat.dk/da/download/file/YXRjX2dyb3Vwcy50eHQ=",
"population_data.txt" = "https://medstat.dk/da/download/file/cG9wdWxhdGlvbl9kYXRhLnR4dA=="
)
seq_atc <- c(1:23)
atc_code_data_list <- link_list_dk[seq_atc]
Data on population sizes of each age group and sex.
age_structure <- read_delim(link_list_dk[["population_data.txt"]], delim = ";", col_names = c(paste0("V", c(1:7))), col_types = cols(V1 = col_character(), V2 = col_character(), V3 = col_character(), V4 = col_character(), V5 = col_character(), V6 = col_character(), V7 = col_character())) %>%
select(year = V1,
region_text = V2,
region = V3,
gender = V4,
age = V5,
denominator_per_year = V6) %>%
mutate(
gender_text = case_when(
gender == "0" ~ "both sexes",
gender == "1" ~ "men",
gender == "2" ~ "women",
T ~ as.character(gender)
)
) %>%
mutate_at(vars(year, age, denominator_per_year), as.numeric) %>%
filter(age %in% 15:84, gender == "2", region == "0") %>%
arrange(year, age, region, gender)
age_structure %>% slice(1:10)
#
#
#
#
#
#
#
#
#
#
#
#
#
Prepare labels for the drugs in English
eng_drug <- read_delim(link_list_dk[["atc_groups.txt"]], delim = ";", col_names = c(paste0("V", c(1:6))), col_types = cols(V1 = col_character(), V2 = col_character(), V3 = col_character(), V4 = col_character(), V5 = col_character(), V6 = col_character())) %>%
filter(V5 == "1") %>%
select(ATC = V1,
drug = V2,
unit_dk = V4)
eng_drug %>% slice(1:10)
#
#
#
#
#
#
#
#
#
#
#
#
#
The medication used in Denmark for either migraine episode relief or migraine prophylaxis:
regex_triptans <- "N02CC$"
regex_ergots <- "N02CA$"
regex_nsaid <- "M01A$"
regex_naproxen <- "M01AE02$"
regex_erenumab <- "N02CX07$|N02CD01$"
regex_galcanezumab <- "N02CX08$|N02CD02$"
regex_fremanezumab <- "N02CD03$"
regex_paracet <- "N02BE01$"
regex_salyc_acid_caff <- "N02BA51$"
regex_metoclopramide <- "A03FA01$"
regex_domperidone <- "A03FA03$"
regex_metoprolol <- "C07AB02$"
regex_propanolol <- "C07AA05$"
regex_tolfenamic <- "M01AG02$"
regex_topiramate <- "N03AX11$"
regex_valproate <- "N03AG01$"
regex_flunarizine <- "N07CA03$"
regex_amitriptyline <- "N06AA09$"
regex_gabapentin <- "N03AX12$"
regex_pizotifen <- "N02CX01$"
regex_lisinopril <- "C09AA03$"
regex_candesartan <- "C09CA06$"
regex_riboflavin <- "A11HA04$"
regex_botulinum <- "M03AX01$"
all_migraine_drugs <- paste(regex_triptans, regex_ergots, regex_nsaid, regex_naproxen, regex_erenumab, regex_galcanezumab, regex_fremanezumab, regex_paracet, regex_salyc_acid_caff, regex_metoclopramide, regex_domperidone, regex_metoprolol, regex_propanolol, regex_tolfenamic, regex_topiramate, regex_valproate, regex_flunarizine, regex_amitriptyline, regex_gabapentin, regex_pizotifen, regex_lisinopril, regex_candesartan, regex_riboflavin, regex_botulinum, sep = "|")
atc_data <- map(atc_code_data_list, ~read_delim(file = .x, delim = ";", trim_ws = T, col_names = c(paste0("V", c(1:14))), col_types = cols(V1 = col_character(), V2 = col_character(), V3 = col_character(), V4 = col_character(), V5 = col_character(),V6 = col_character(), V7 = col_character(), V8 = col_character(), V9 = col_character(), V10 = col_character(), V11 = col_character(), V12 = col_character(), V13 = col_character(), V14 = col_character()))) %>%
bind_rows()
atc_data %<>%
rename(
ATC = V1,
year = V2,
sector = V3,
region = V4,
gender = V5,
age = V6,
number_of_people = V7,
patients_per_1000_inhabitants = V8,
turnover = V9,
regional_grant_paid = V10,
quantity_sold_1000_units = V11,
quantity_sold_units_per_unit_1000_inhabitants_per_day = V12,
percentage_of_sales_in_the_primary_sector = V13
) %>%
mutate(
year = as.numeric(year),
region_text = case_when(
region == "0" ~ "DK",
region == "1" ~ "Region Hovedstaden",
region == "2" ~ "Region Midtjylland",
region == "3" ~ "Region Nordjylland",
region == "4" ~ "Region Sjælland",
region == "5" ~ "Region Syddanmark",
T ~ NA_character_
),
gender_text = case_when(
gender == "0" ~ "both sexes",
gender == "1" ~ "men",
gender == "2" ~ "women",
T ~ as.character(gender)
)
) %>%
mutate_at(vars(turnover, regional_grant_paid, quantity_sold_1000_units, quantity_sold_units_per_unit_1000_inhabitants_per_day, number_of_people, patients_per_1000_inhabitants), as.numeric) %>%
select(-V14) %>%
filter(
gender == "2",
region == "0",
sector =="0"
) %>%
filter(str_detect(ATC, all_migraine_drugs)) %>%
mutate(
age = case_when(
age == "95+" ~ "95",
T ~ as.character(age)
),
age = as.integer(age)
) %>%
filter(age %in% 15:84) %>%
mutate(
age_cat = case_when(
age %in% 15:24 ~ "15-24",
age %in% 25:34 ~ "25-34",
age %in% 35:44 ~ "35-44",
age %in% 45:54 ~ "45-54",
age %in% 55:64 ~ "55-64",
age %in% 65:74 ~ "65-74",
age %in% 75:84 ~ "75-84",
T ~ NA_character_
)
) %>%
left_join(age_structure) %>%
left_join(eng_drug) %>%
group_by(ATC, year, age_cat) %>%
mutate(
numerator = sum(number_of_people),
denominator = sum(denominator_per_year),
patients_per_1000_inhabitants = numerator / denominator * 1000
) %>%
filter(row_number() == 1) %>%
ungroup() %>%
select(year, ATC, drug, unit_dk, gender, age_cat, patients_per_1000_inhabitants) %>%
mutate(
country = "Denmark"
)
atc_data %>% slice(1:10)
#
#
#
#
#
#
#
#
#
#
#
#
#
Checking if all medication utilization was available
atc_data %>%
distinct(ATC, drug) %>%
filter(is.na(drug))
#
#
#
#
atc_data %<>% mutate(
drug = case_when(
ATC == "N02CD01" ~ "Erenumab",
T ~ drug
)
)
Results: Denmark
All results for Denmark
results_dk <- atc_data %>%
filter((age_cat == "25-34" | age_cat == "35-44"), year == "2019") %>%
select(ATC, drug, age_cat, patients_per_1000_inhabitants, unit_dk, country)
Swedish data
Now, to Swedish data. The full guide on how to upload aggregated Swedish medication data can be found
here. I have the data loaded and will use the same list of medications as for the Danish data.
Keep only the drugs used in migraine, population of women between 18 and 84 on the national level
data_se %<>%
filter(gender == "Kvinnor", region == "Riket") %>%
filter(str_detect(string = ATC, pattern = all_migraine_drugs))
Swedish data is grouped by age categories:
data_se %>% distinct(age_group)
#
#
#
#
#
#
#
#
#
#
#
#
#
#
#
#
#
#
#
#
#
#
Categorize age groups according to Danish data:
data_se %<>% filter(! age_group %in% c("0-4", "5-9", "10-14", "85+", "Totalt")) %>%
mutate(
age_cat = case_when(
age_group == "15-19" | age_group == "20-24" ~ "15-24",
age_group == "25-29" | age_group == "30-34" ~ "25-34",
age_group == "35-39" | age_group == "40-44" ~ "35-44",
age_group == "45-49" | age_group == "50-54" ~ "45-54",
age_group == "55-59" | age_group == "60-64" ~ "55-64",
age_group == "65-69" | age_group == "70-74" ~ "65-74",
age_group == "75-79" | age_group == "80-84" ~ "75-84",
T ~ "other"
)
) %>%
group_by(ATC, year, age_cat) %>%
mutate(
numerator = sum(`Antal patienter`),
denominator = sum(Befolkning),
patients_per_1000_inhabitants = numerator / denominator * 1000
) %>%
filter(row_number() == 1) %>%
ungroup() %>%
select(year, ATC, gender, age_cat, patients_per_1000_inhabitants) %>%
mutate(
country = "Sweden"
)
data_se %>% count(age_cat)
#
#
#
#
#
#
#
#
#
#
Add English labels to the ATC codes in Swedish data
data_se %<>%
left_join(eng_drug)
Check if all medication utilization was available
data_se %>%
distinct(ATC, drug) %>%
filter(is.na(drug))
#
#
#
#
#
#
data_se %<>% mutate(
drug = case_when(
ATC == "N02CD01" ~ "Erenumab",
ATC == "N02CD03" ~ "Galcanezumab",
ATC == "N02CX07" ~ "Erenumab",
T ~ drug
)
)
Join Danish and Swedish data
atc_data %<>% full_join(data_se)
plot_utilization <- function(data, drug_regex, title, max_year){
data %>%
filter(!is.na(patients_per_1000_inhabitants)) %>%
mutate(label = if_else(year == max_year, as.character(age_cat), NA_character_),
value = case_when(
age_cat == "25-34" ~"#3E9286",
age_cat == "35-44" ~ "#E8B88A",
T ~ "gray"
)) %>%
filter(str_detect(ATC, {{ drug_regex }})) %>%
ggplot(aes(x = year, y = patients_per_1000_inhabitants, group = age_cat, color = value)) +
geom_line() +
facet_grid(rows = vars(drug, ATC), cols = vars(country), scales = "free", drop = F) +
theme_light(base_size = 14) +
scale_x_continuous(breaks = c(seq(1995, 2019, 4))) +
scale_color_identity() +
theme(plot.caption = element_text(hjust = 0, size = 10),
legend.position = "none",
panel.spacing = unit(0.8, "cm")) +
labs(y = "Female patients\nper 1,000 women in the population", title = paste0(title, " utilization in women"), subtitle = "aged between 15 and 84 years", caption = "dataviz by Elena Dudukina Twitter @evpatora") +
ggrepel::geom_label_repel(aes(label = label), na.rm = TRUE, nudge_x = 3, hjust = 0.5, direction = "y", segment.size = 0.1, segment.colour = "black", show.legend = F)
}
list_regex <- list(regex_triptans, regex_ergots, regex_nsaid, regex_naproxen, regex_erenumab, regex_paracet, regex_salyc_acid_caff, regex_metoclopramide, regex_domperidone, regex_metoprolol, regex_propanolol, regex_topiramate, regex_valproate, regex_flunarizine, regex_amitriptyline, regex_gabapentin, regex_pizotifen, regex_lisinopril, regex_candesartan, regex_botulinum, regex_tolfenamic)
list_title <- list("triptans", "ergots", "NSAIDs", "naproxen", "erenumab", "paracetamol", "salycilic acid and caffeine", "metoclopramide", "domperidone", "metoprolol", "propanolol", "topiramate", "valproate", "flunarizine", "amitriptyline", "gabapentin", "pizotifen", "lisinopril", "candesartan", "botulinum", "tolfenamic") %>%
map_if(. != "NSAIDs", ~ str_to_sentence(.))
list_plots <- pmap(list(list_regex, list_title), ~plot_utilization(data = atc_data, drug_regex = ..1, title = ..2, max_year = 2019)) %>%
setNames(., c("triptans", "ergots", "NSAIDs", "naproxen", "erenumab", "paracetamol", "salycilic acid and caffeine", "metoclopramide", "domperidone", "metoprolol", "propanolol", "topiramate", "valproate", "flunarizine", "amitriptyline", "gabapentin", "pizotifen", "lisinopril", "candesartan", "botulinum", "tolfenamic"))
Combined results for Denmark and Sweden
results_all <- data_se %>%
filter((age_cat == "25-34" | age_cat == "35-44"), year == "2019", !is.na(patients_per_1000_inhabitants)) %>%
select(ATC, drug, age_cat, patients_per_1000_inhabitants, country) %>%
full_join(results_dk) %>%
select(-unit_dk) %>%
arrange(ATC, age_cat) %>%
pivot_wider(names_from = country, values_from = patients_per_1000_inhabitants) %>%
relocate(Sweden, .after = Denmark)
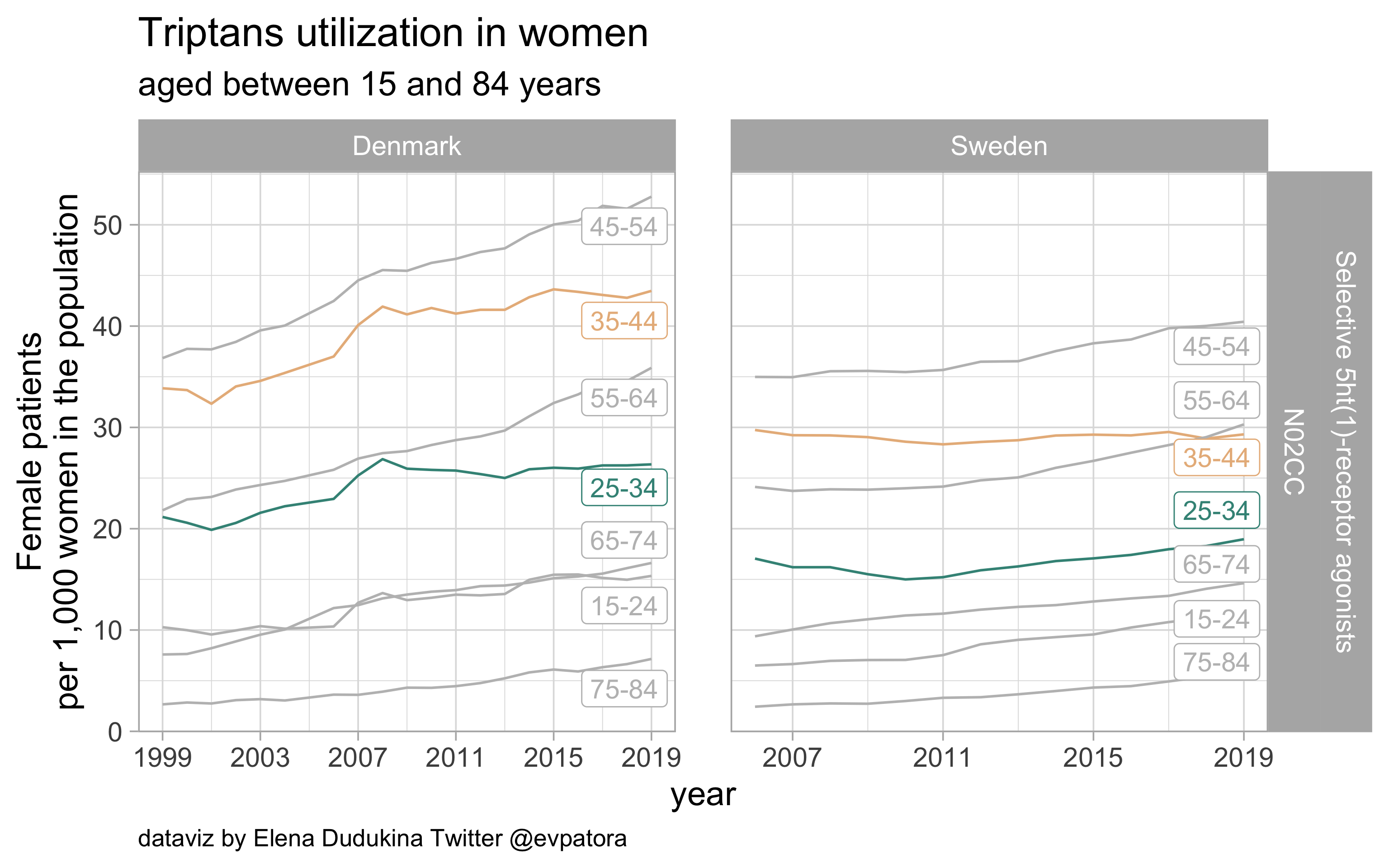
Triptans
- Acute migraine treatment
- Danish women of ages between 25 and 34 utilize triptans at a higher rate (26 women per 1000 females in this age group) than women in the same age group in Sweden (19 women per 1000 females)
- In the age category 35-44 years, Danish women utilize triptans at 1.5 times higher rate
list_plots$triptans
results_all %>%
filter(str_detect(ATC, regex_triptans)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N02CC |
Selective 5ht(1)-receptor agonists |
25-34 |
26.35166 |
18.96388 |
| N02CC |
Selective 5ht(1)-receptor agonists |
35-44 |
43.47541 |
29.31439 |
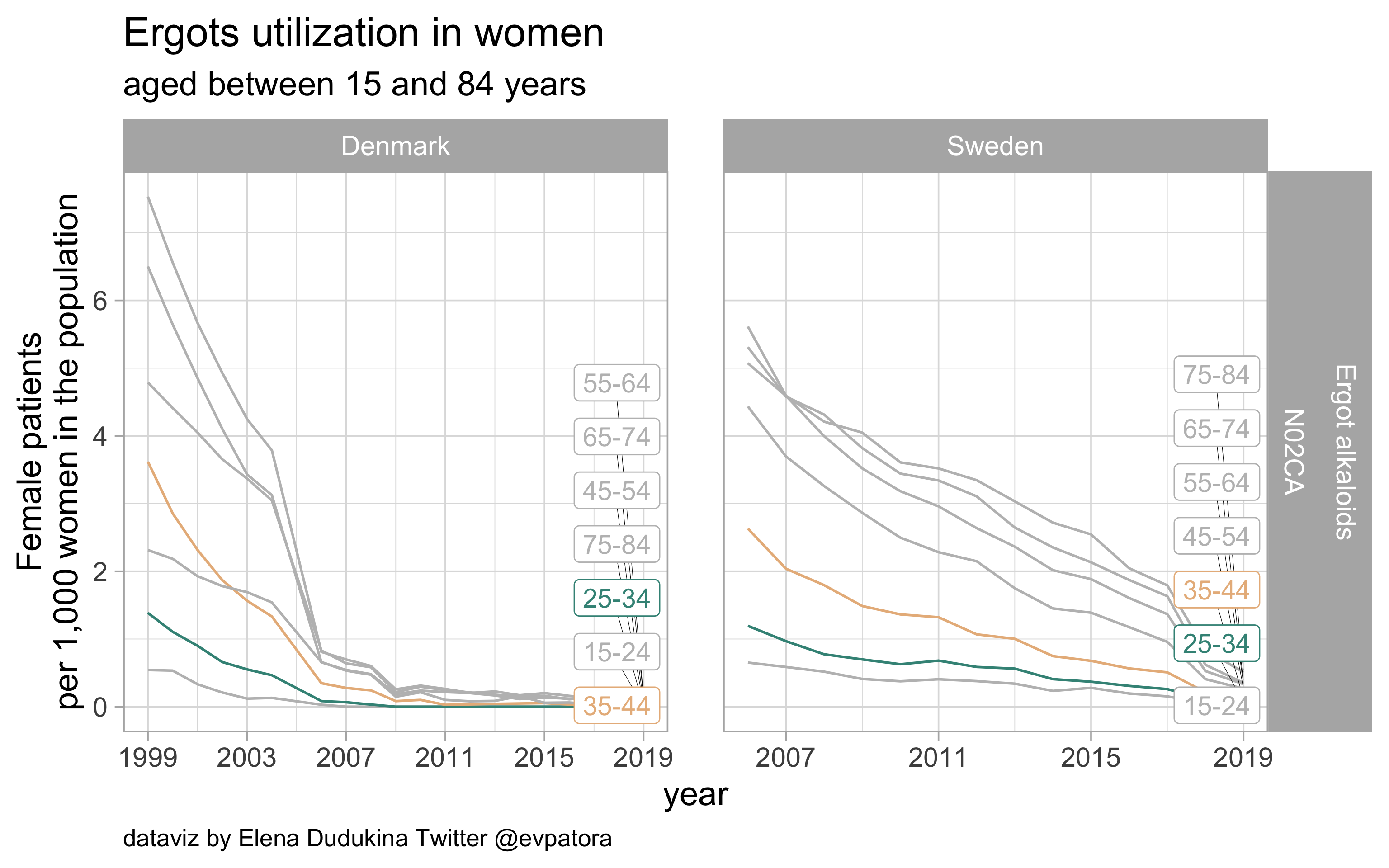
Ergots
- Acute migraine treatment
- Utilization is very low in Sweden and is zero in Denmark (in 2019)
list_plots$ergots
results_all %>%
filter(str_detect(ATC, regex_ergots)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N02CA |
Ergot alkaloids |
25-34 |
0 |
0.0560297 |
| N02CA |
Ergot alkaloids |
35-44 |
0 |
0.1105720 |
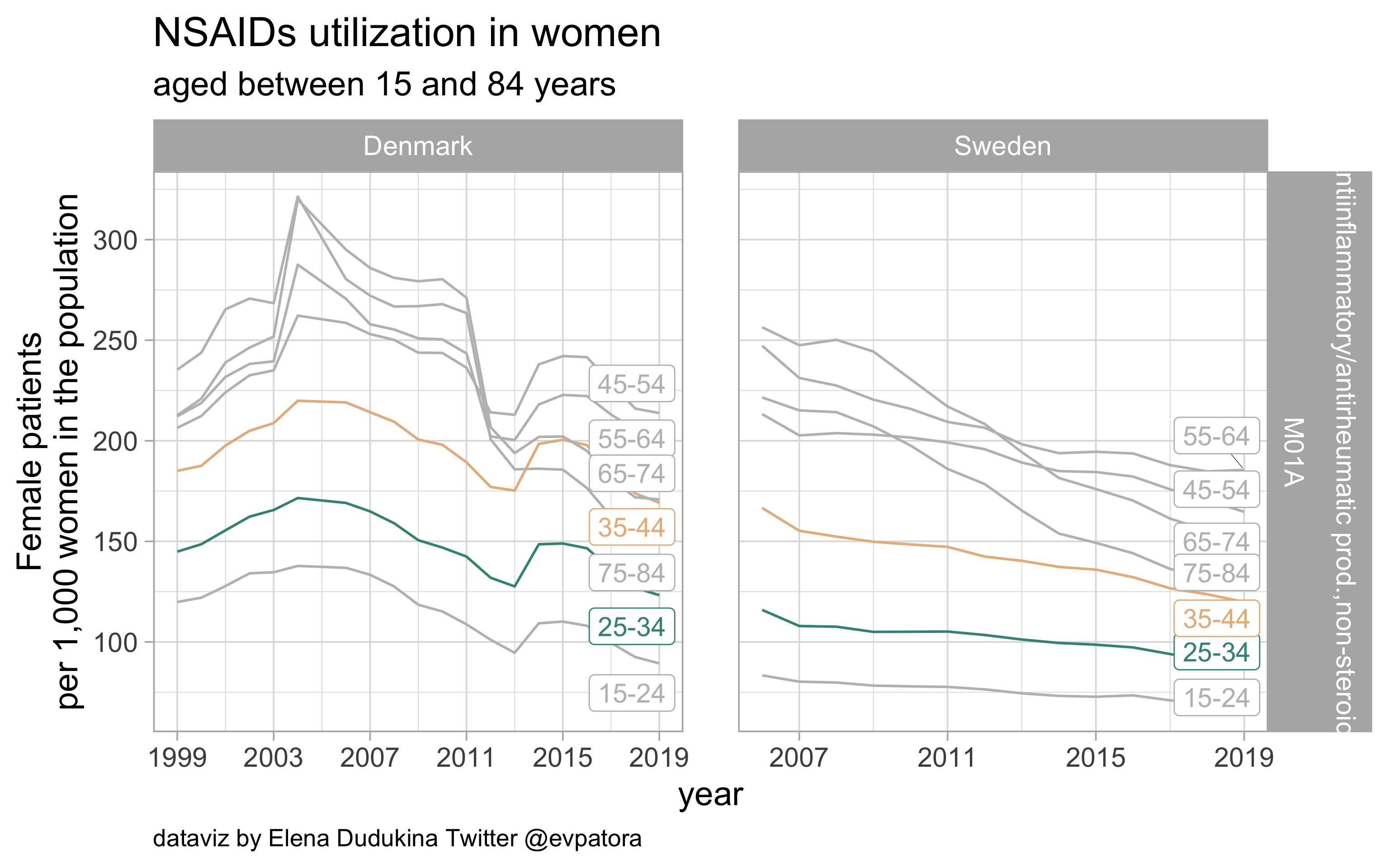
NSAIDs
- Non‐specific acute treatment: indication for aggregated data is unknown
- Danish women of both age categories of interest (25-34 years and 35-44 years) utilize NSAIDs at a higher rate than Swedish women
list_plots$NSAIDs
results_all %>%
filter(str_detect(ATC, regex_nsaid)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| M01A |
Antiinflammatory/antirheumatic prod.,non-steroids |
25-34 |
123.2309 |
87.11605 |
| M01A |
Antiinflammatory/antirheumatic prod.,non-steroids |
35-44 |
169.2165 |
119.87122 |
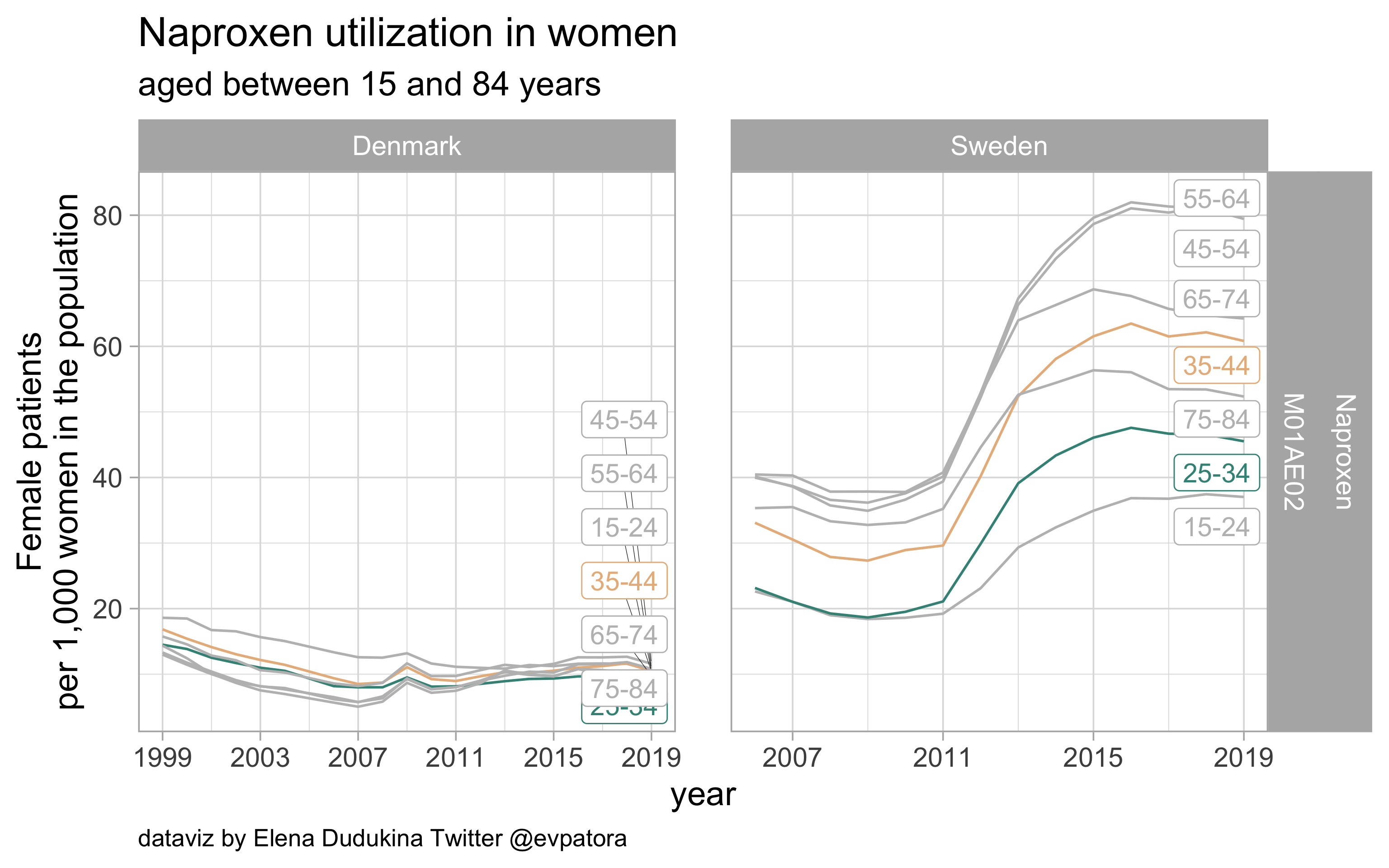
Naproxen
- Non‐specific acute treatment: indication for aggregated data is unknown
- Swedish women of both age categories of interest (25-34 years and 35-44 years) utilize naproxen at a higher rate than Danish women
list_plots$naproxen
results_all %>%
filter(str_detect(ATC, regex_naproxen)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| M01AE02 |
Naproxen |
25-34 |
8.890147 |
45.52050 |
| M01AE02 |
Naproxen |
35-44 |
10.451094 |
60.82099 |
Erenumab
- Prophylactic treatment
- No record found in Danish data
- Very low utilization according to Swedish data
list_plots$erenumab
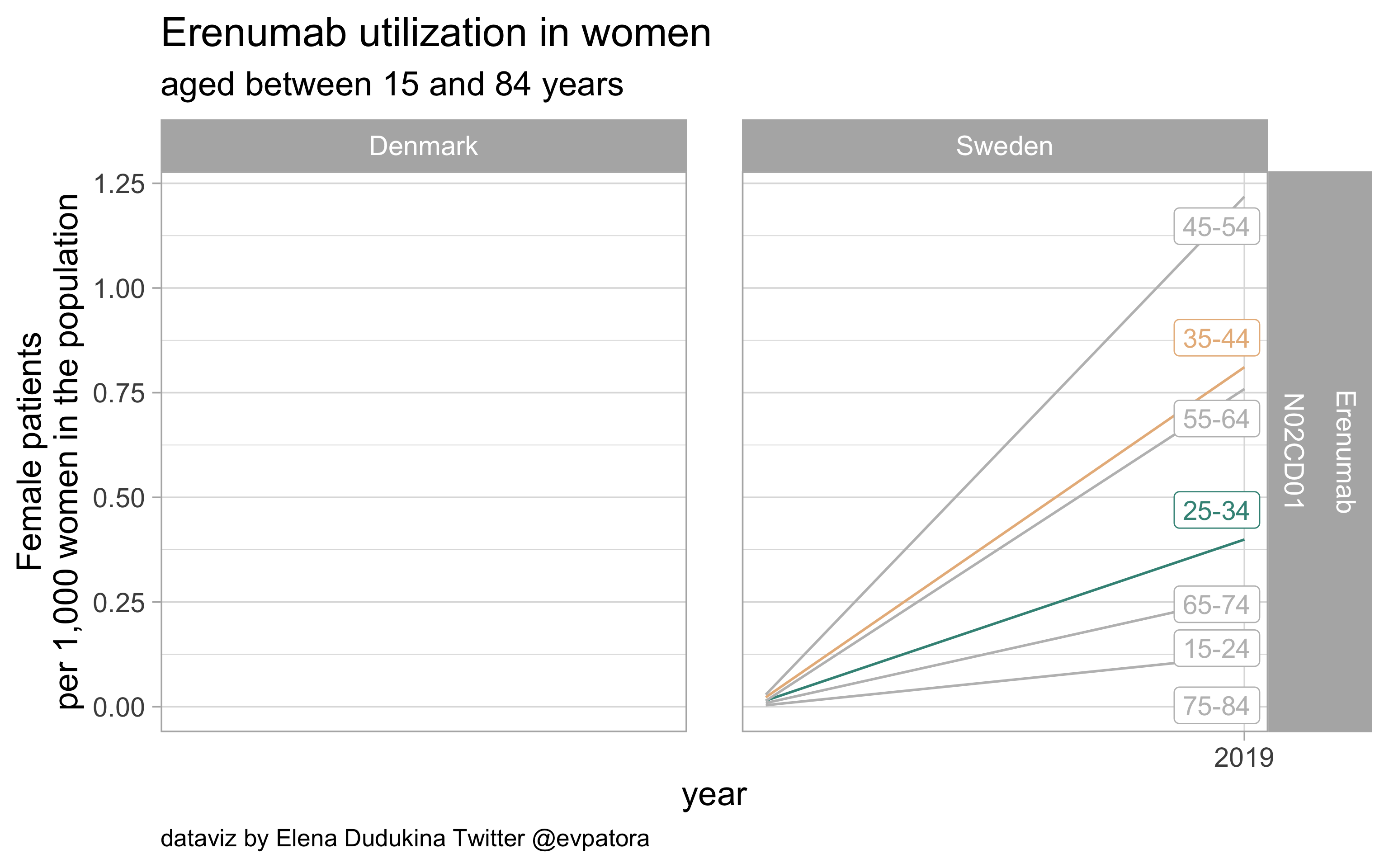
results_all %>%
filter(str_detect(ATC, regex_erenumab)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N02CD01 |
Erenumab |
25-34 |
NA |
0.3993909 |
| N02CD01 |
Erenumab |
35-44 |
NA |
0.8108611 |
Paracetamol
- Nonspecific acute treatment: indication for aggregated data is unknown
- Utilization rate of paracetamol (via prescription) in Denmark is remarkably higher than in Sweden for women in age categories 25-34 and 35-44 years
list_plots$paracetamol
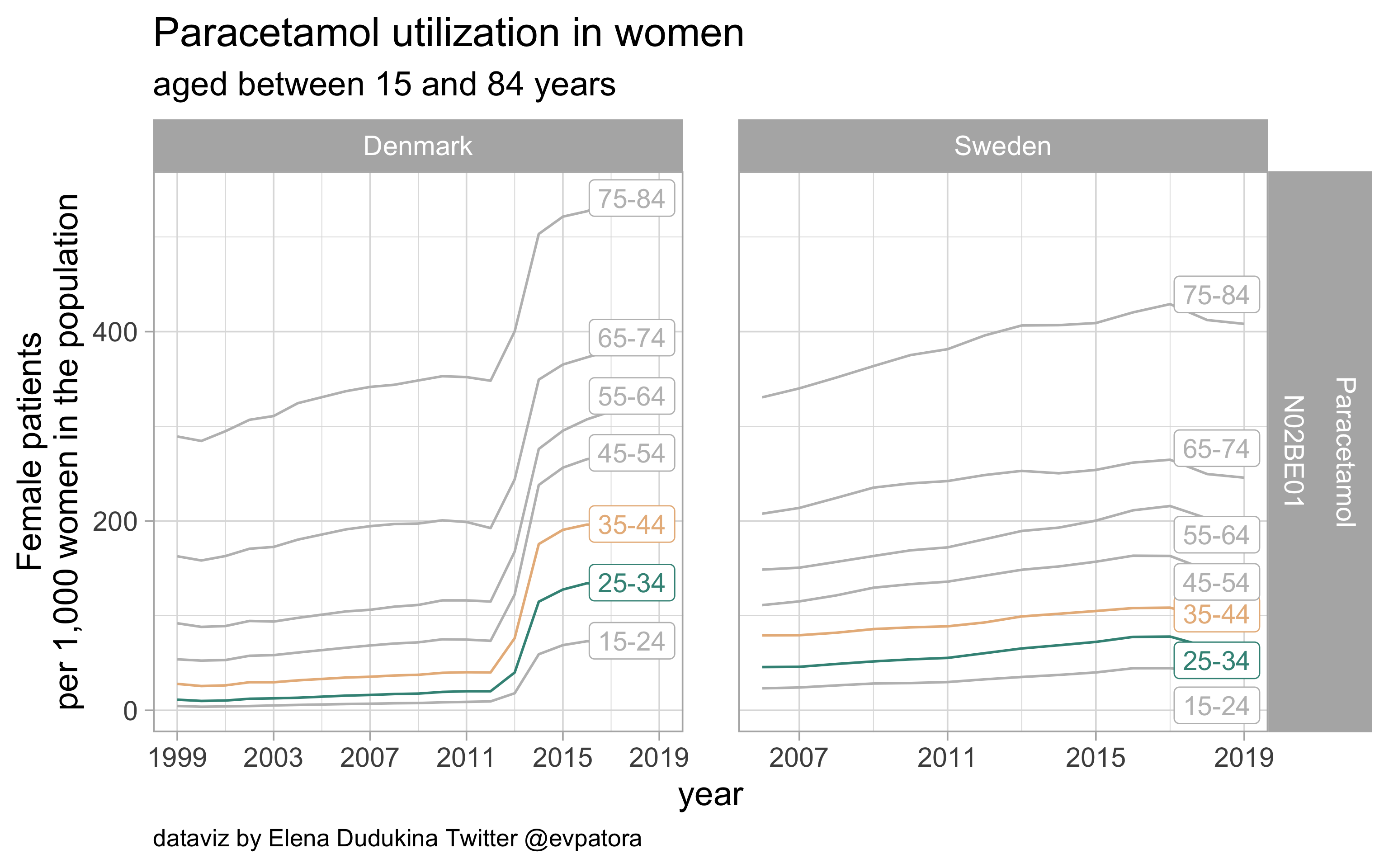
results_all %>%
filter(str_detect(ATC, regex_paracet)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N02BE01 |
Paracetamol |
25-34 |
134.8802 |
64.02322 |
| N02BE01 |
Paracetamol |
35-44 |
196.8056 |
93.76663 |
Salycilic acid and caffeine
- Nonspecific acute treatment: indication for aggregated data is unknown
- Low utilization in both Sweden and Denmark; the utilization rate among Danish women is 8 times that among Swedish women aged 25-34 and 11 times that among Swedish women aged 35-44
list_plots$`salycilic acid and caffeine`
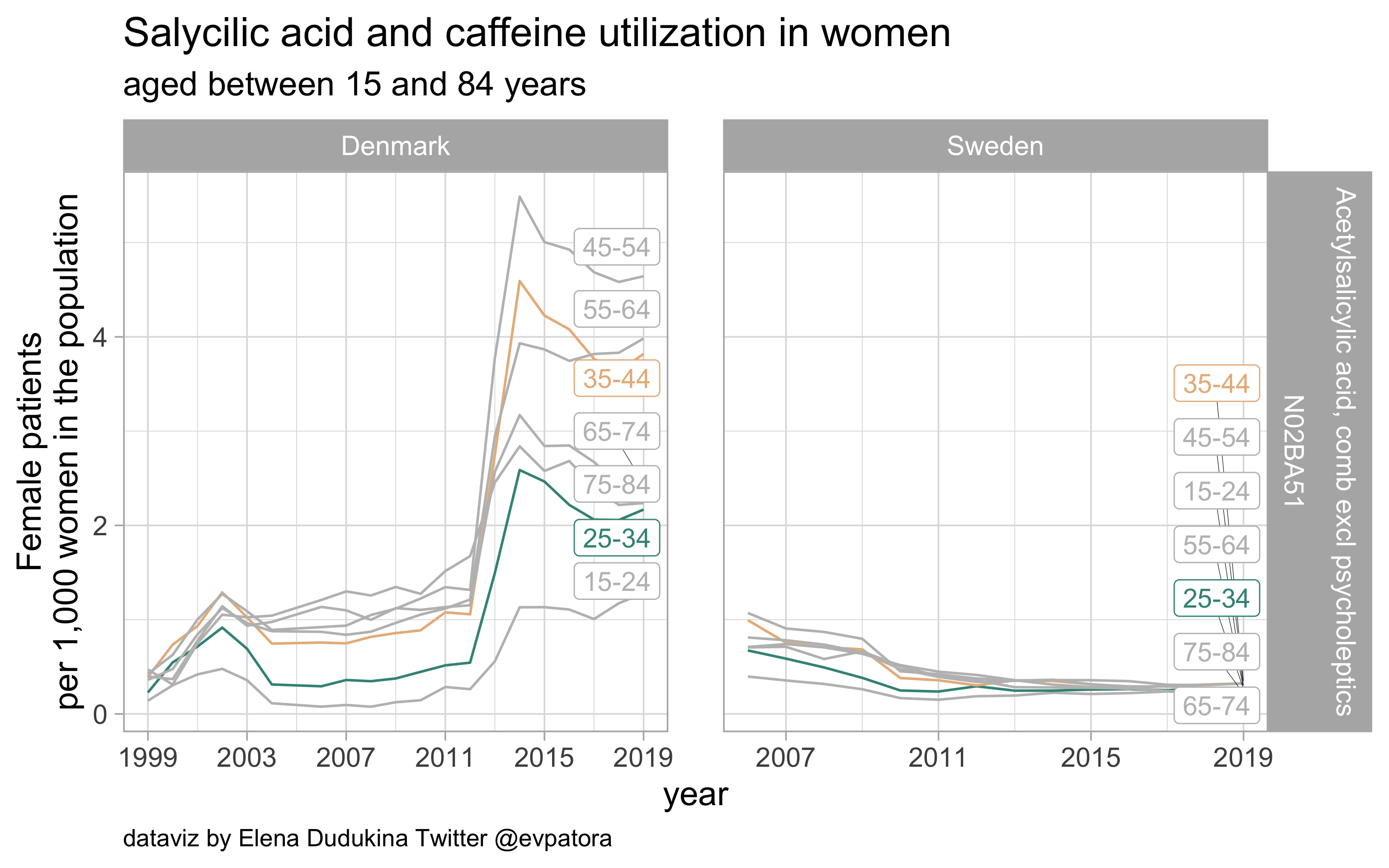
results_all %>%
filter(str_detect(ATC, regex_salyc_acid_caff)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N02BA51 |
Acetylsalicylic acid, comb excl psycholeptics |
25-34 |
2.168261 |
0.2629084 |
| N02BA51 |
Acetylsalicylic acid, comb excl psycholeptics |
35-44 |
3.818285 |
0.3253059 |
Metoclopramide
- Nonspecific acute treatment: indication for aggregated data is unknown
- Utilization rates are similar in Denmark and Sweden among women of age categories of interest
list_plots$metoclopramide
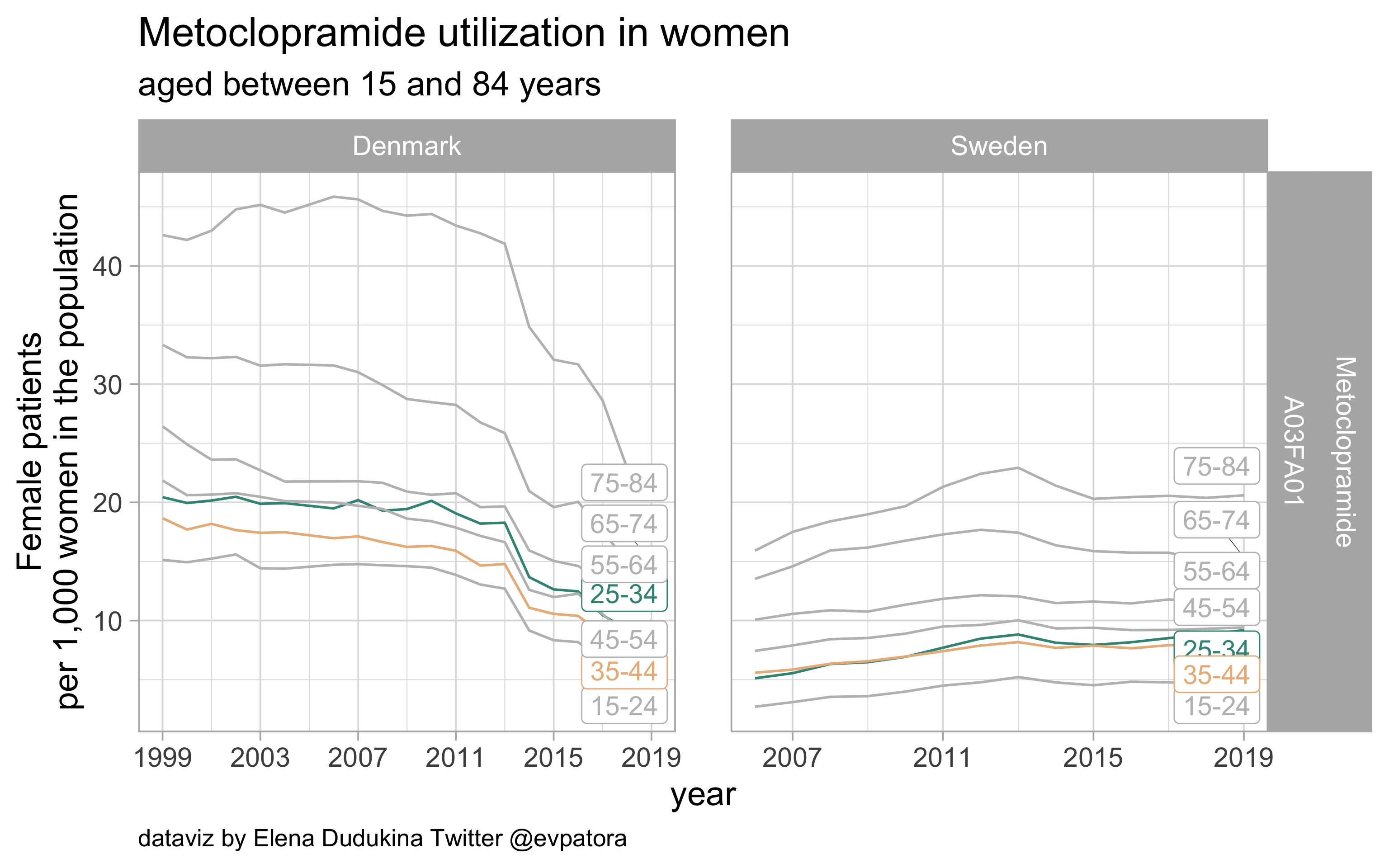
results_all %>%
filter(str_detect(ATC, regex_metoclopramide)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| A03FA01 |
Metoclopramide |
25-34 |
9.967407 |
9.190300 |
| A03FA01 |
Metoclopramide |
35-44 |
7.405591 |
8.401867 |
Domperidone
- Nonspecific acute treatment: indication for aggregated data is unknown
- Low utilization in Denmark and Sweden; although higher rates among Danish women than in Swedish women
list_plots$domperidone
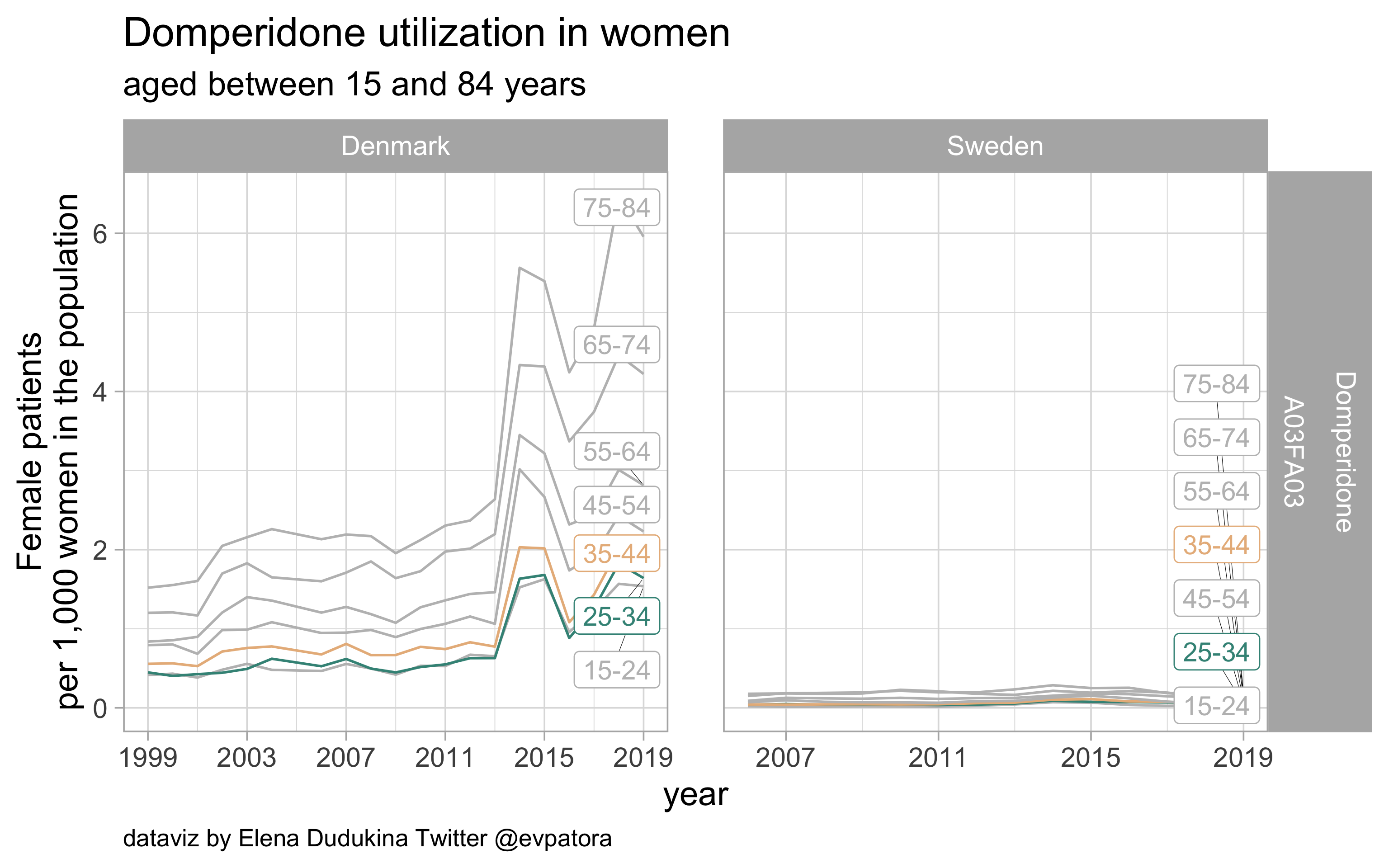
results_all %>%
filter(str_detect(ATC, regex_domperidone)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| A03FA03 |
Domperidone |
25-34 |
1.640624 |
0.0416631 |
| A03FA03 |
Domperidone |
35-44 |
1.748027 |
0.0689072 |
Metoprolol
- Nonspecific prophylactic treatment: indication for aggregated data is unknown
- Utilization rate of metoprolol is higher among Swedish women between 25 and 44 years than among Danish women in the same age band
list_plots$metoprolol
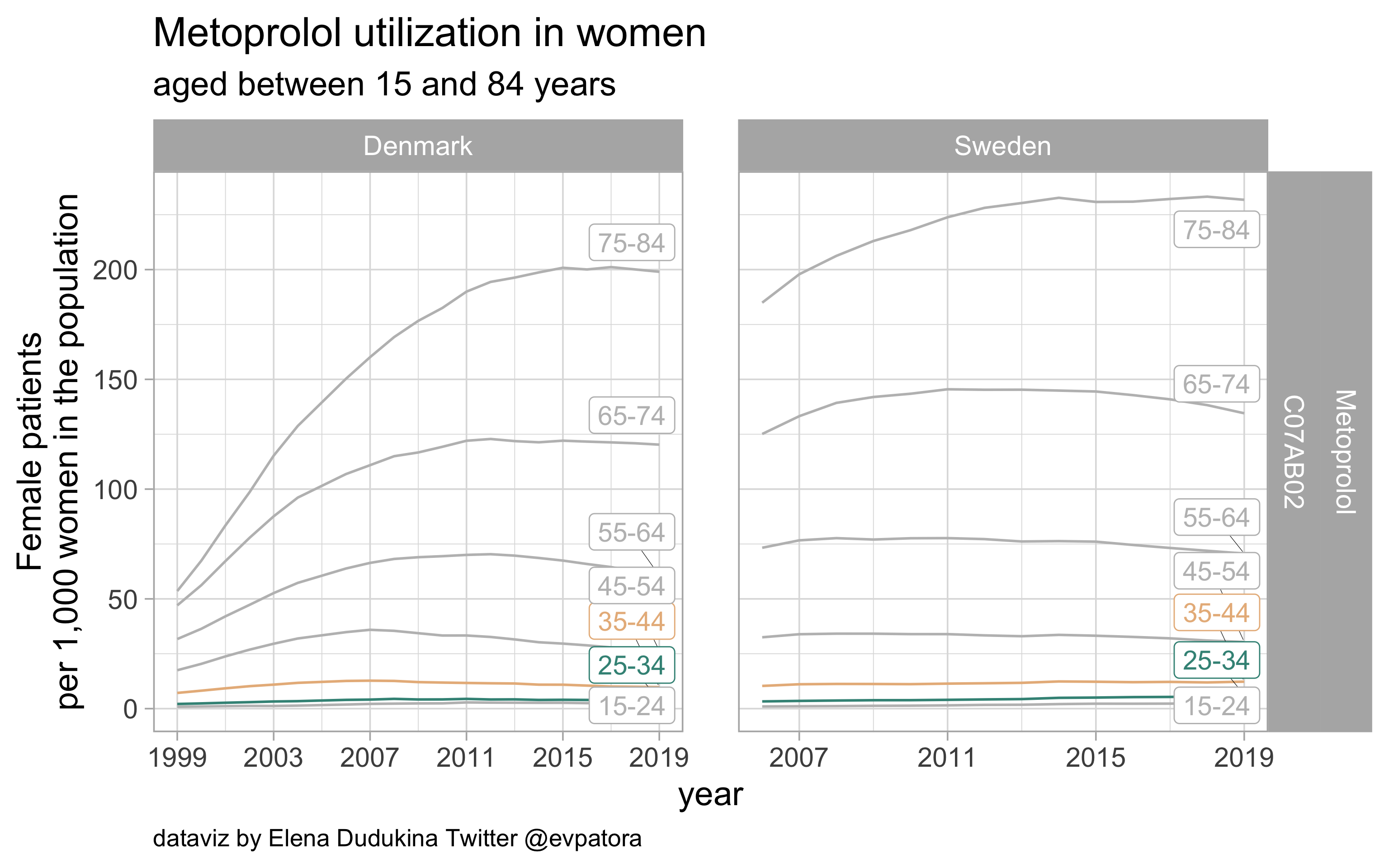
results_all %>%
filter(str_detect(ATC, regex_metoprolol)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| C07AB02 |
Metoprolol |
25-34 |
3.712701 |
6.271011 |
| C07AB02 |
Metoprolol |
35-44 |
9.832298 |
12.291115 |
Propranolol
- Nonspecific prophylactic treatment: indication for aggregated data is unknown
- Utilization rate of metoprolol is higher among Swedish women between 25 and 44 years than among Danish women in the same age band
list_plots$propanolol
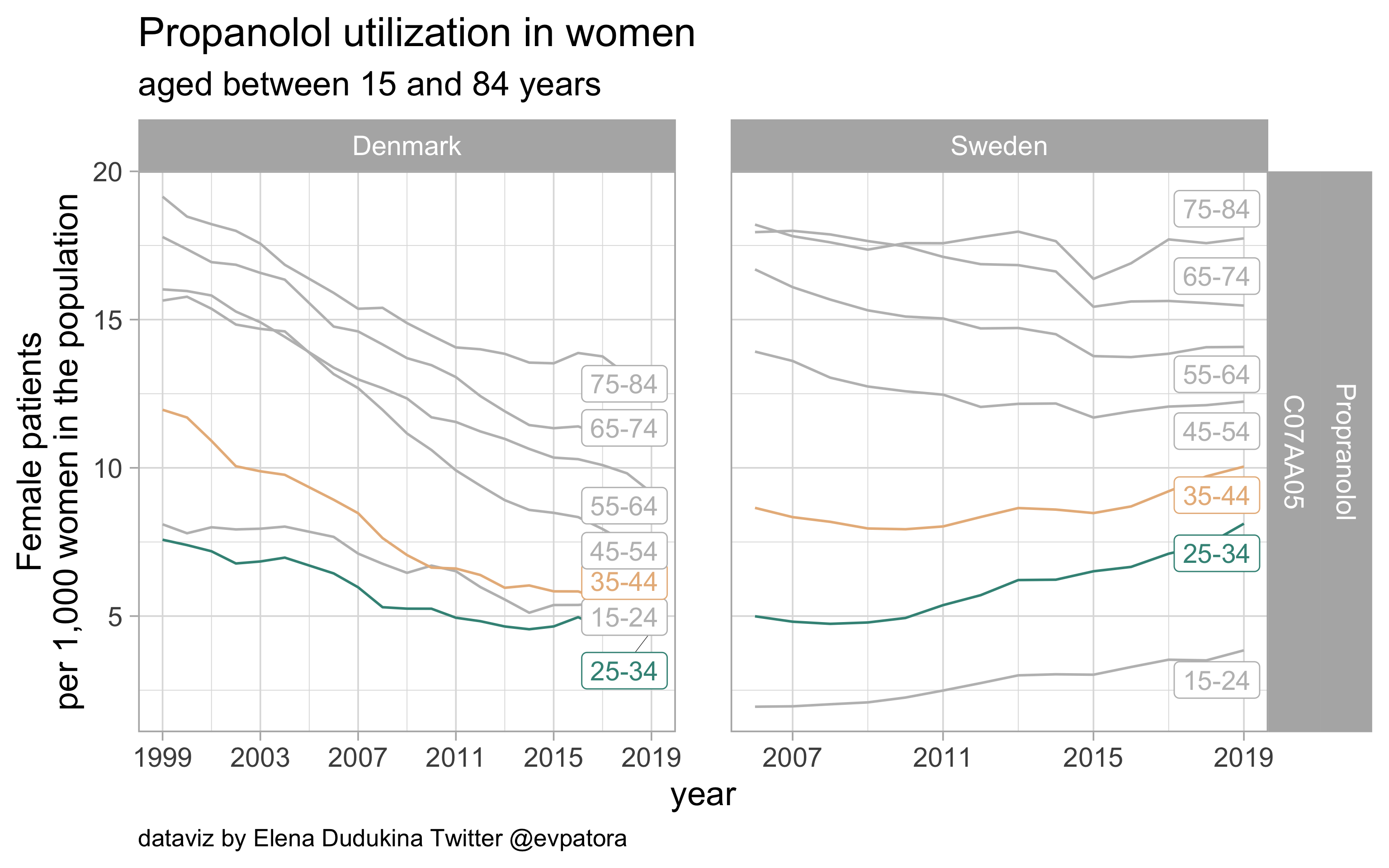
results_all %>%
filter(str_detect(ATC, regex_propanolol)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| C07AA05 |
Propranolol |
25-34 |
4.495914 |
8.117116 |
| C07AA05 |
Propranolol |
35-44 |
5.235527 |
10.042819 |
Tolfenamic acid
- Nonspecific acute treatment: indication for aggregated data is unknown
- No available data in Sweden; low utilization in Denmark
list_plots$tolfenamic
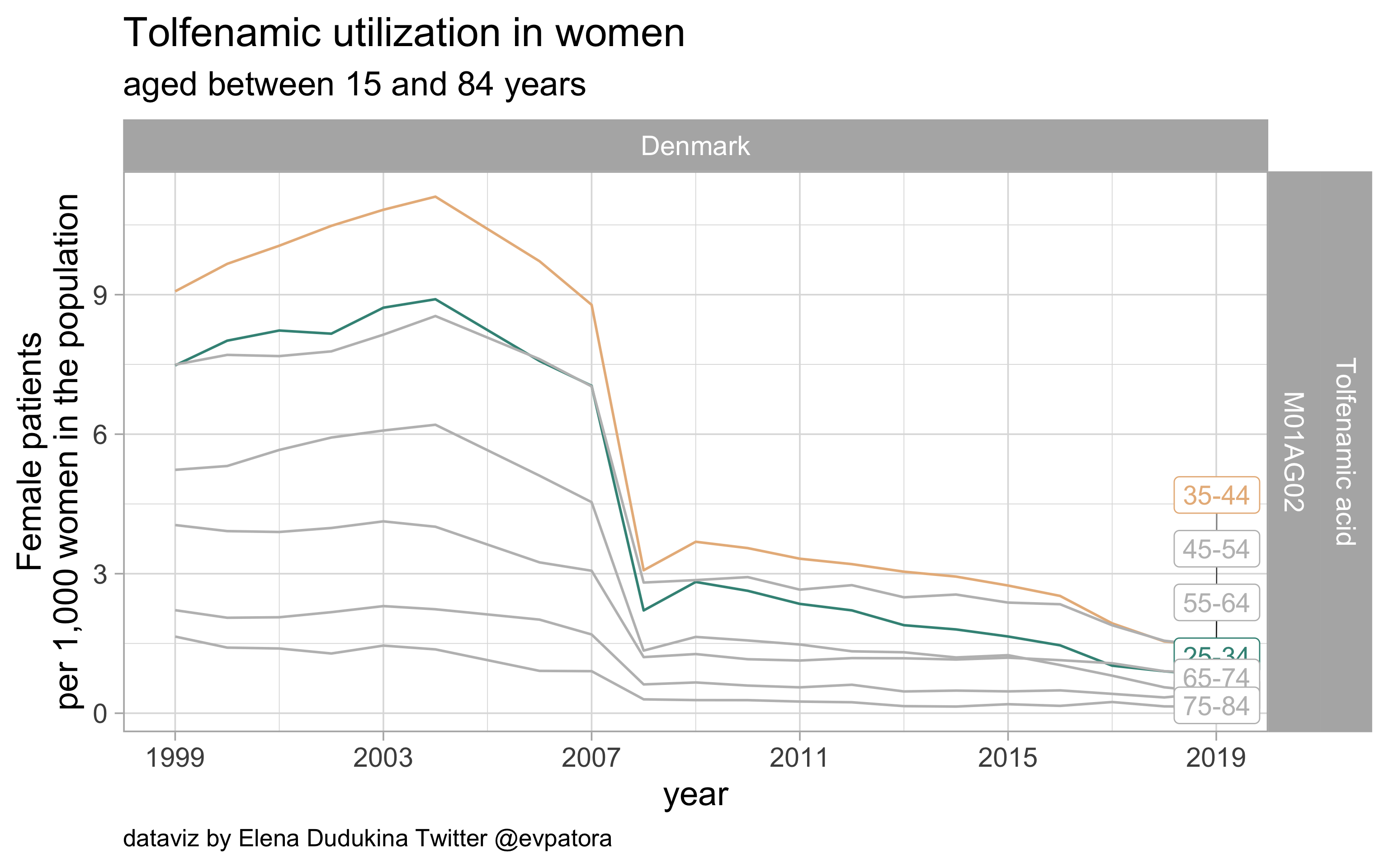
results_all %>%
filter(str_detect(ATC, regex_tolfenamic)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| M01AG02 |
Tolfenamic acid |
25-34 |
0.8106935 |
NA |
| M01AG02 |
Tolfenamic acid |
35-44 |
1.4571648 |
NA |
Topiramate
- Prophylactic migraine treatment
- Indication is unknown
- Similar utilization among women in the age categories 25-34 and 35-44 years in Sweden and Denmark
list_plots$topiramate
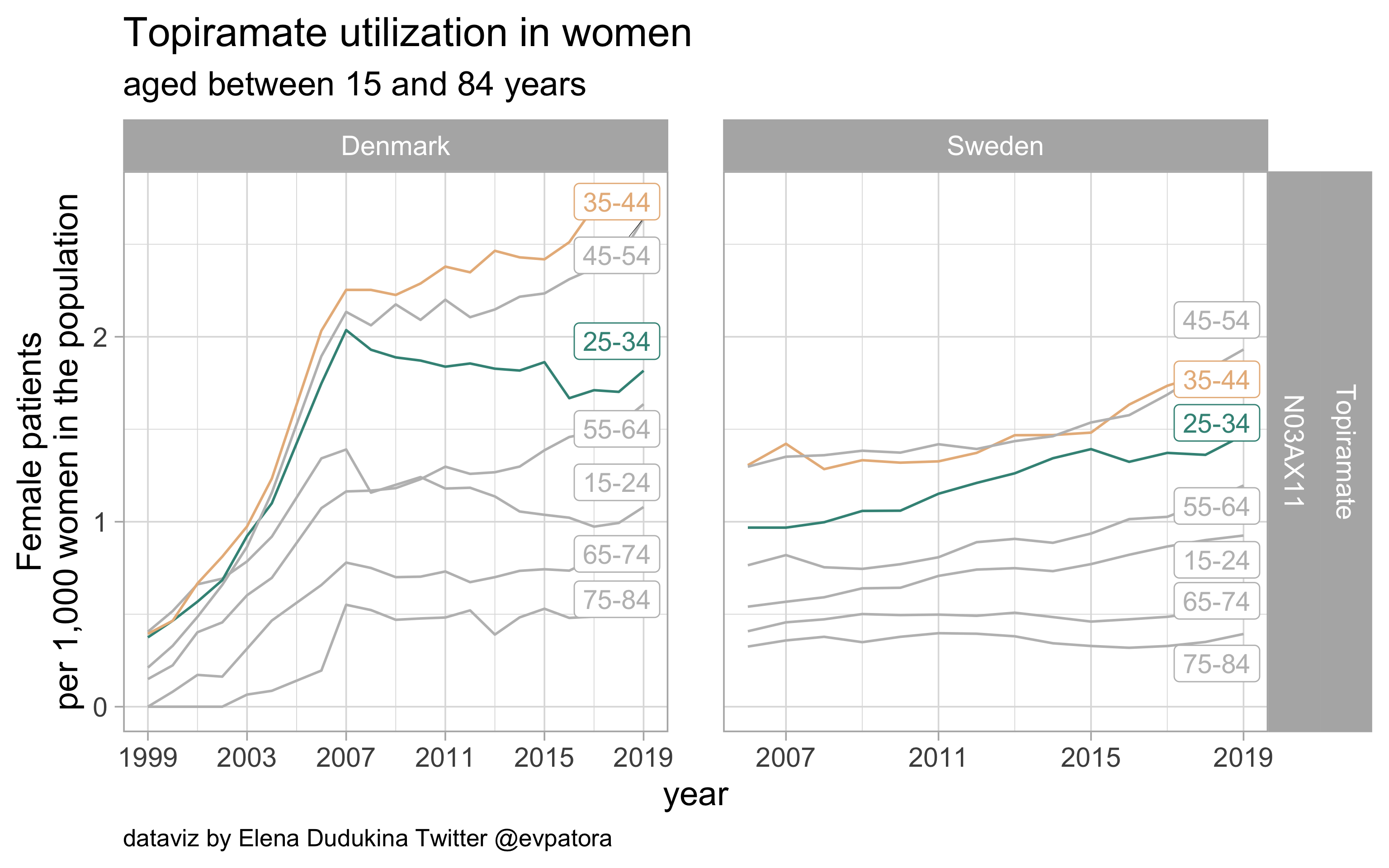
results_all %>%
filter(str_detect(ATC, regex_topiramate)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N03AX11 |
Topiramate |
25-34 |
1.816503 |
1.468264 |
| N03AX11 |
Topiramate |
35-44 |
2.757492 |
1.836456 |
Valproate
- Prophylactic migraine treatment
- Indication is unknown
- Similar utilization among women in the age categories 25-34 and 35-44 years in Sweden and Denmark
list_plots$valproate
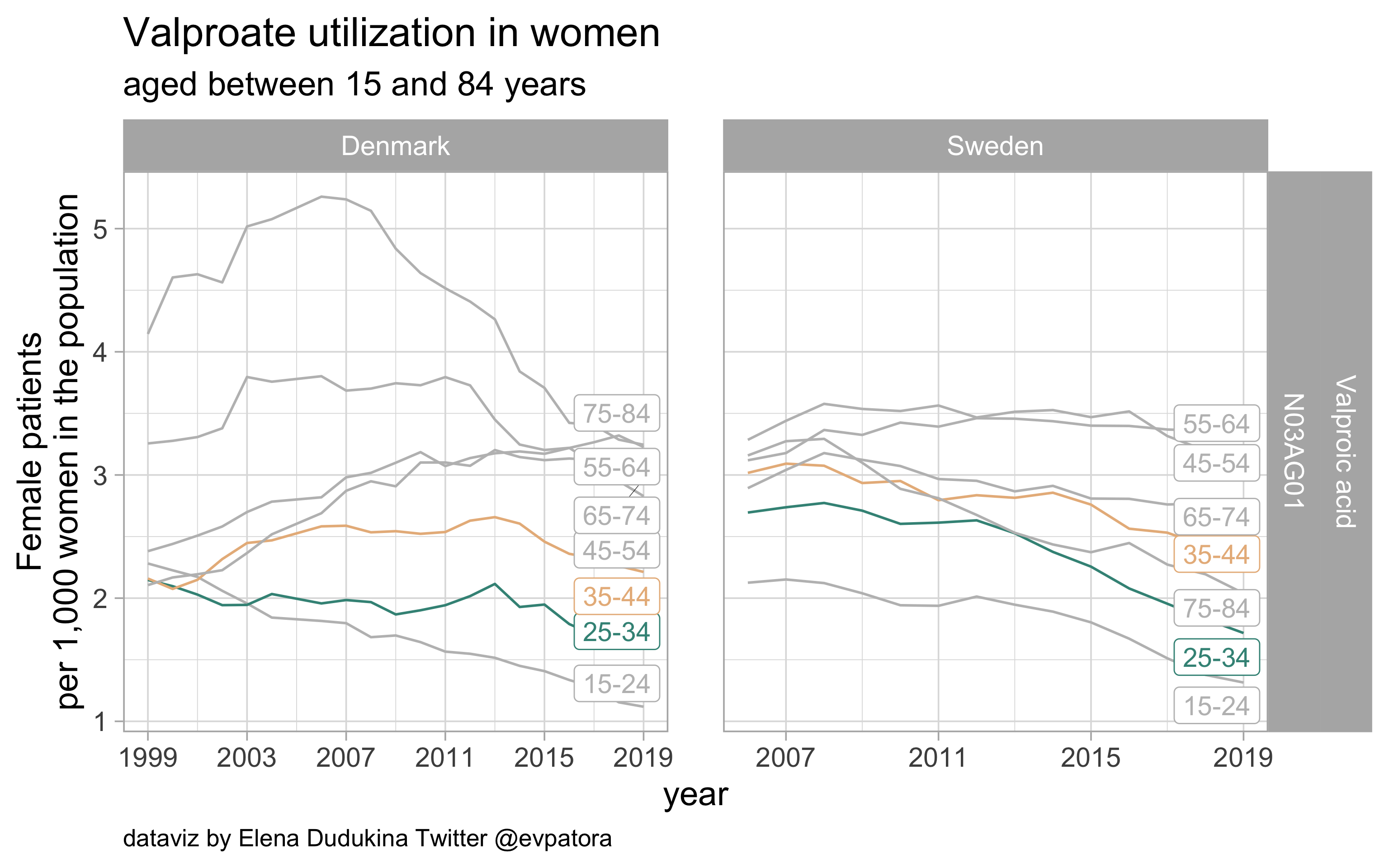
results_all %>%
filter(str_detect(ATC, regex_valproate)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N03AG01 |
Valproic acid |
25-34 |
1.588410 |
1.716806 |
| N03AG01 |
Valproic acid |
35-44 |
2.212837 |
2.222657 |
Flunarizin
- Prophylactic migraine treatment
- Very low utilization in Denmark and Sweden
list_plots$flunarizine
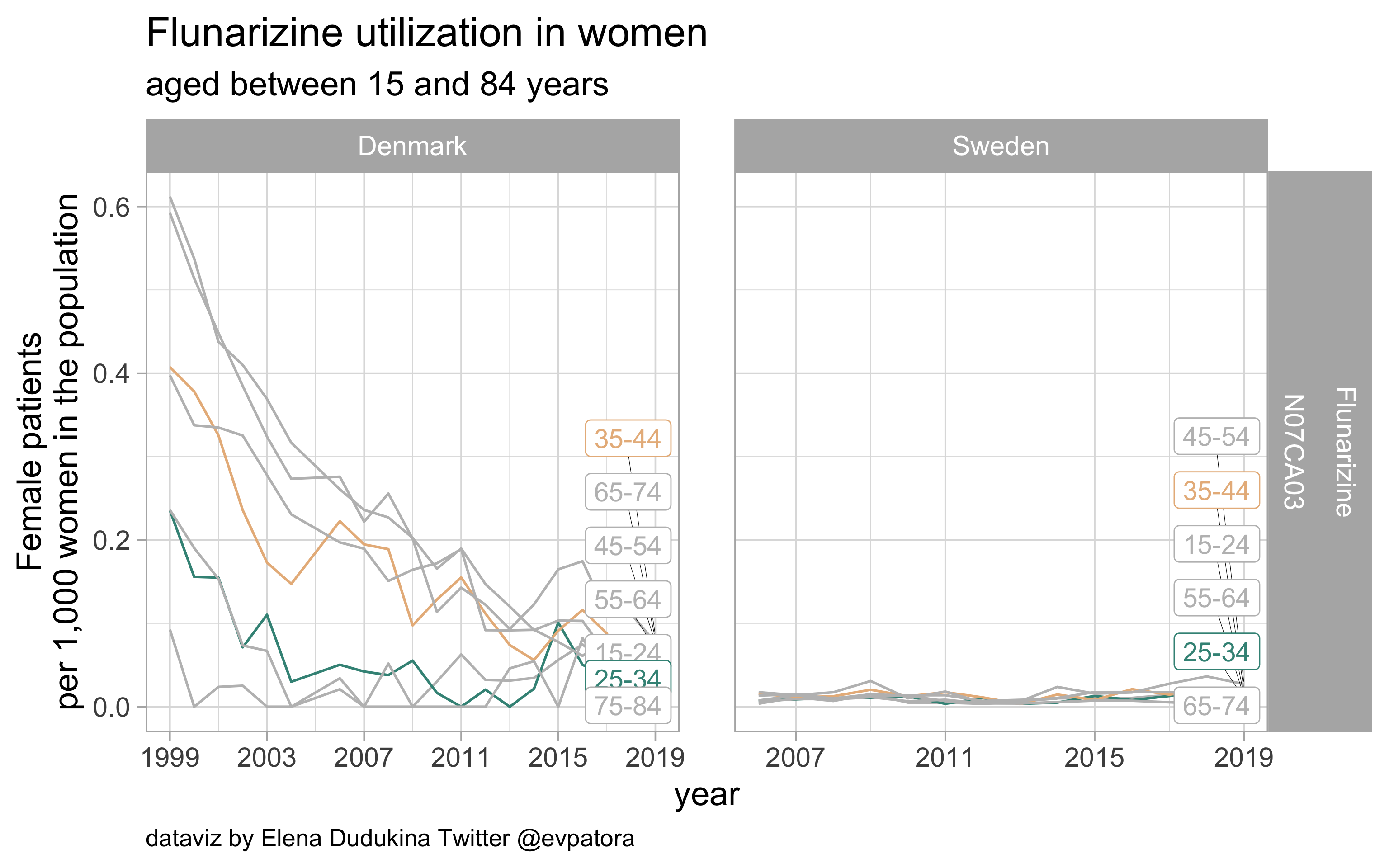
results_all %>%
filter(str_detect(ATC, regex_flunarizine)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N07CA03 |
Flunarizine |
25-34 |
0.0154212 |
0.0100566 |
| N07CA03 |
Flunarizine |
35-44 |
0.0854206 |
0.0192299 |
Amitriptyline
- Prophylactic migraine treatment
- Indication is unknown
- Utilization rate is higher among Swedish women in the age categories 25-34 and 35-44 years
list_plots$amitriptyline
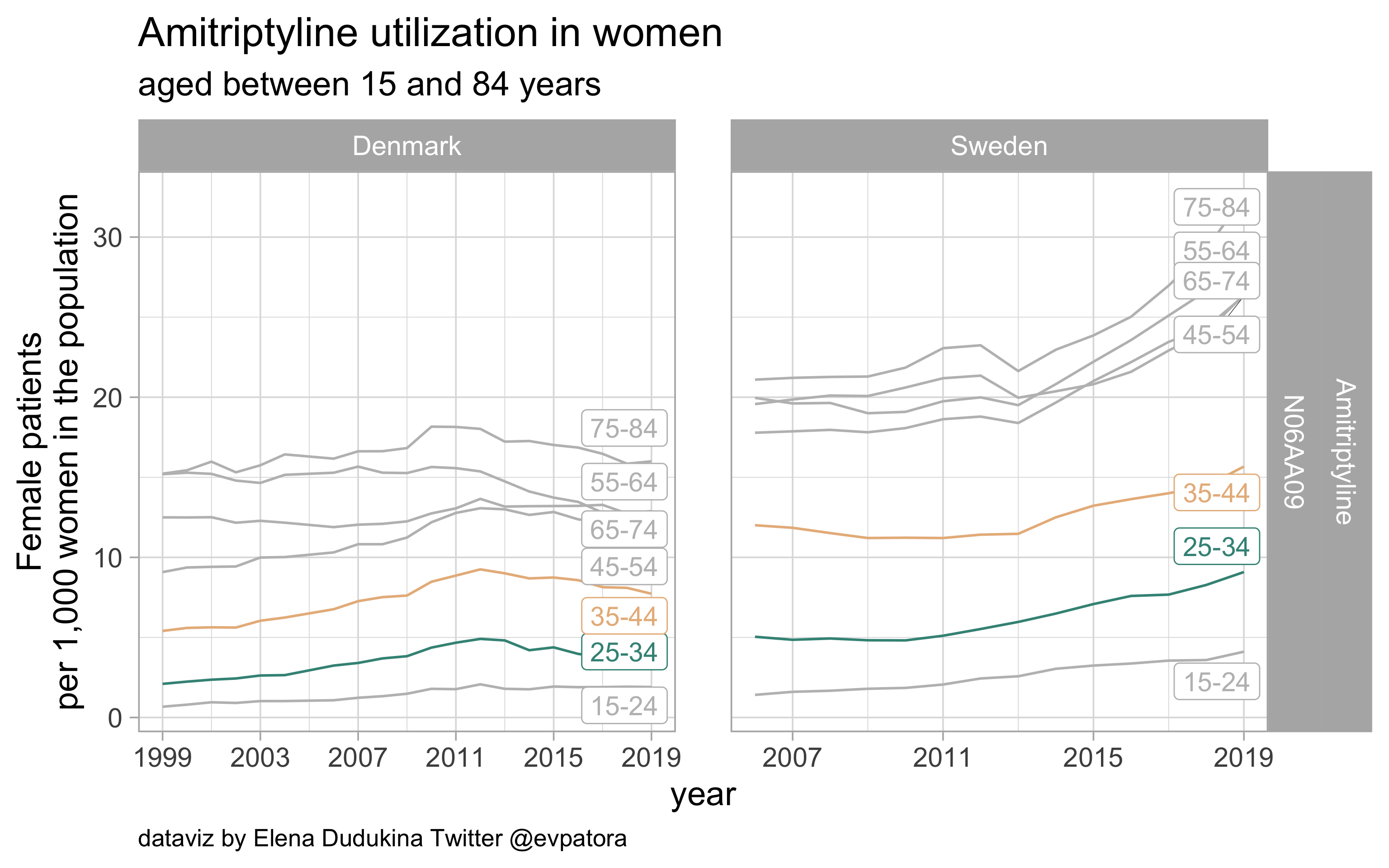
results_all %>%
filter(str_detect(ATC, regex_amitriptyline)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N06AA09 |
Amitriptyline |
25-34 |
3.822626 |
9.08255 |
| N06AA09 |
Amitriptyline |
35-44 |
7.727821 |
15.67237 |
Gabapentin
- Prophylactic migraine treatment
- Indication is unknown
- Higher utilization rate among Danish women in the age categories 25-34 and 35-44 years compared with Swedish women of the same age categories
list_plots$gabapentin
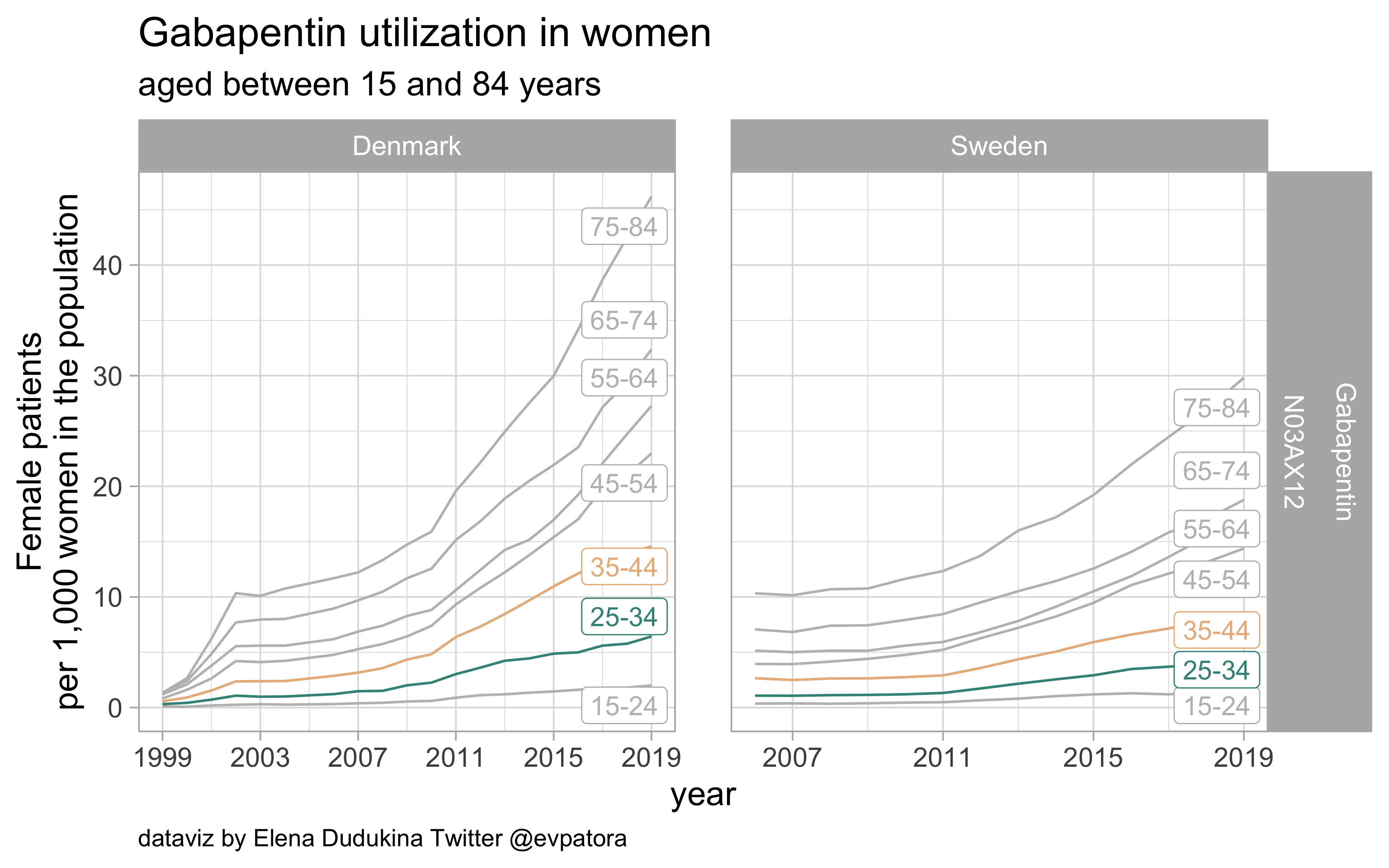
results_all %>%
filter(str_detect(ATC, regex_gabapentin)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N03AX12 |
Gabapentin |
25-34 |
6.427837 |
4.344453 |
| N03AX12 |
Gabapentin |
35-44 |
14.563093 |
8.613396 |
Pizotifen
- Prophylactic migraine treatment
- Very low utilization among Swedish women and zero among Danish women in the age categories 25-34 and 35-44 years
list_plots$pizotifen
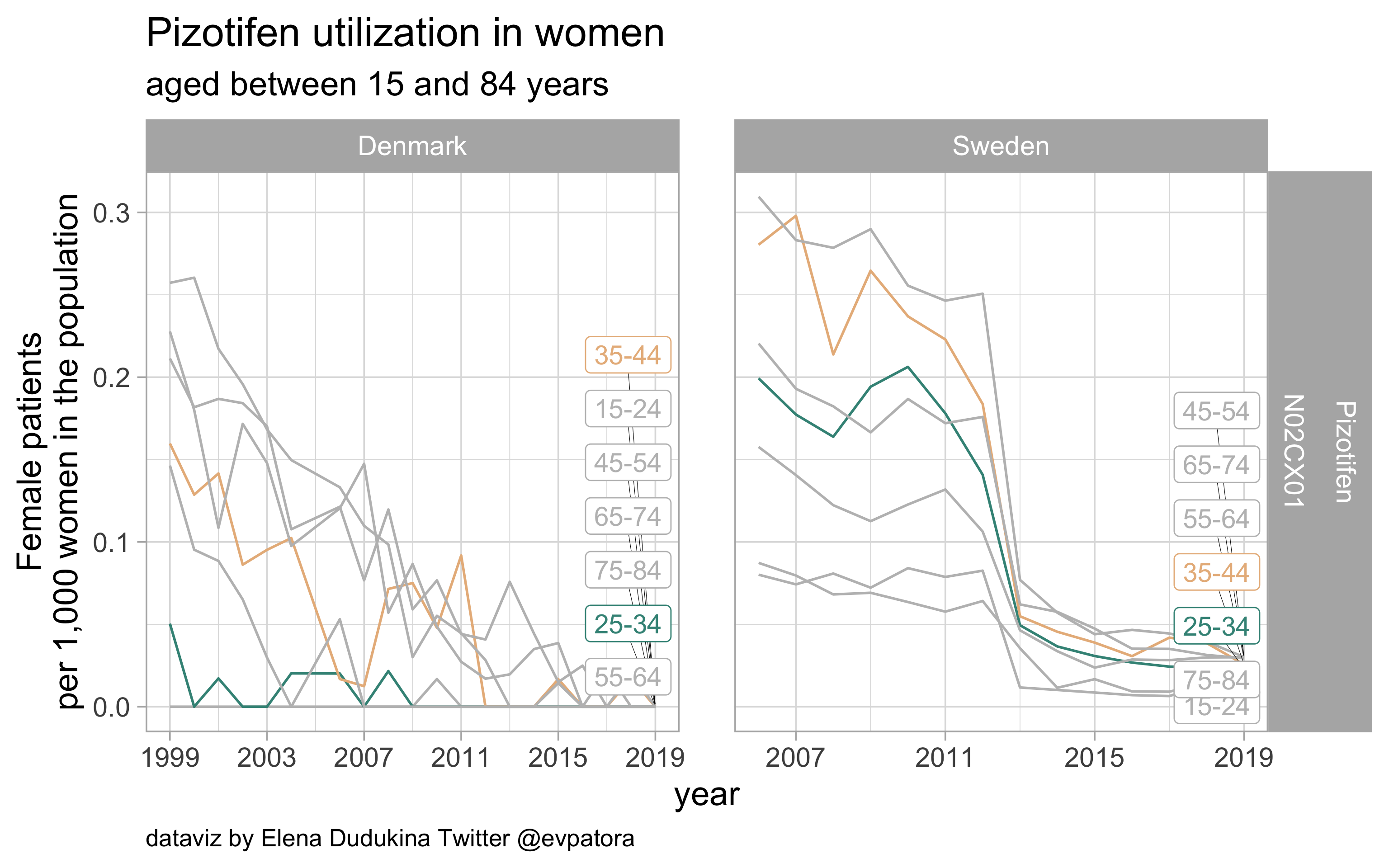
results_all %>%
filter(str_detect(ATC, regex_pizotifen)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| N02CX01 |
Pizotifen |
25-34 |
0 |
0.0229865 |
| N02CX01 |
Pizotifen |
35-44 |
0 |
0.0256399 |
Lisinopril
- Prophylactic migraine treatment
- Very low utilization among Swedish women and nearly zero among Danish women in the age categories 25-34 and 35-44 years
list_plots$lisinopril
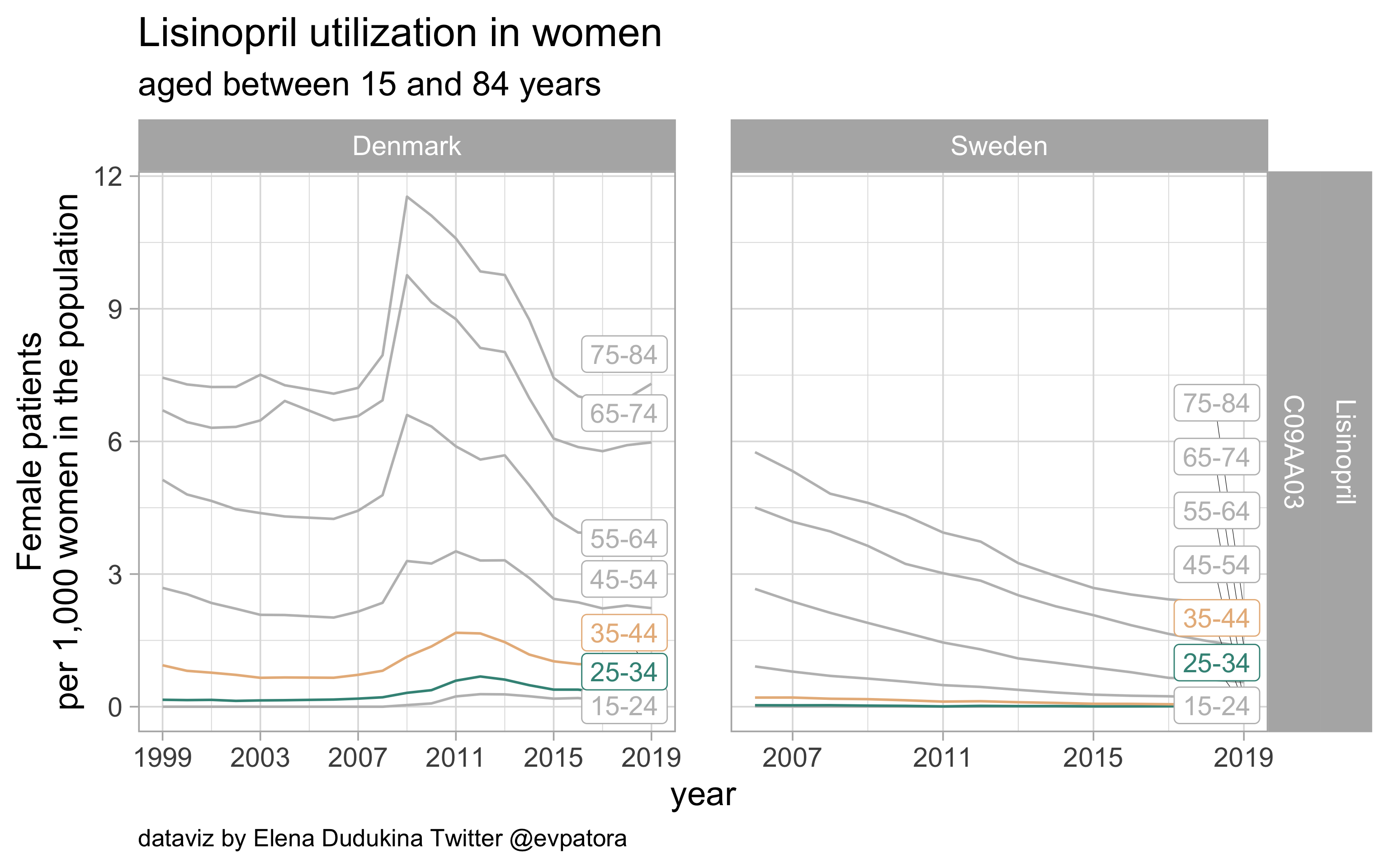
results_all %>%
filter(str_detect(ATC, regex_lisinopril)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| C09AA03 |
Lisinopril |
25-34 |
0.2967962 |
0.0086199 |
| C09AA03 |
Lisinopril |
35-44 |
0.8212592 |
0.0512798 |
Candesartan
- Prophylactic migraine treatment
- Indication is unknown
- Similar utilization rates among Swedish and Danish women in the age categories 25-34 and 35-44 years
list_plots$candesartan
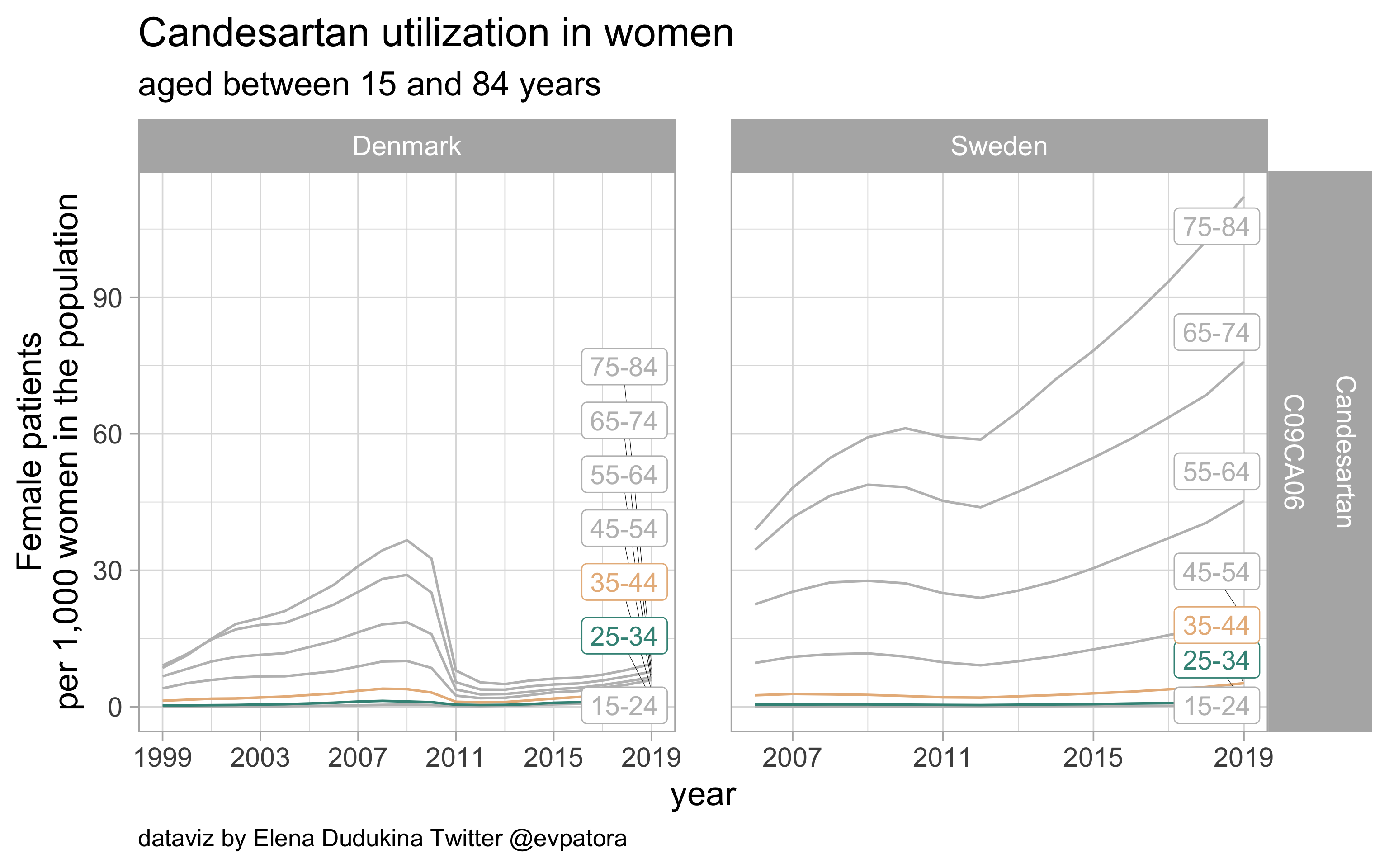
results_all %>%
filter(str_detect(ATC, regex_candesartan)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| C09CA06 |
Candesartan |
25-34 |
1.728563 |
1.282935 |
| C09CA06 |
Candesartan |
35-44 |
3.727034 |
5.190472 |
Botulinum toxin
- Prophylactic migraine treatment
- Indication is unknown
- Higher utilization rates among Swedish women than among Danish women (nearly zero) in the age categories 25-34 and 35-44 years
list_plots$botulinum
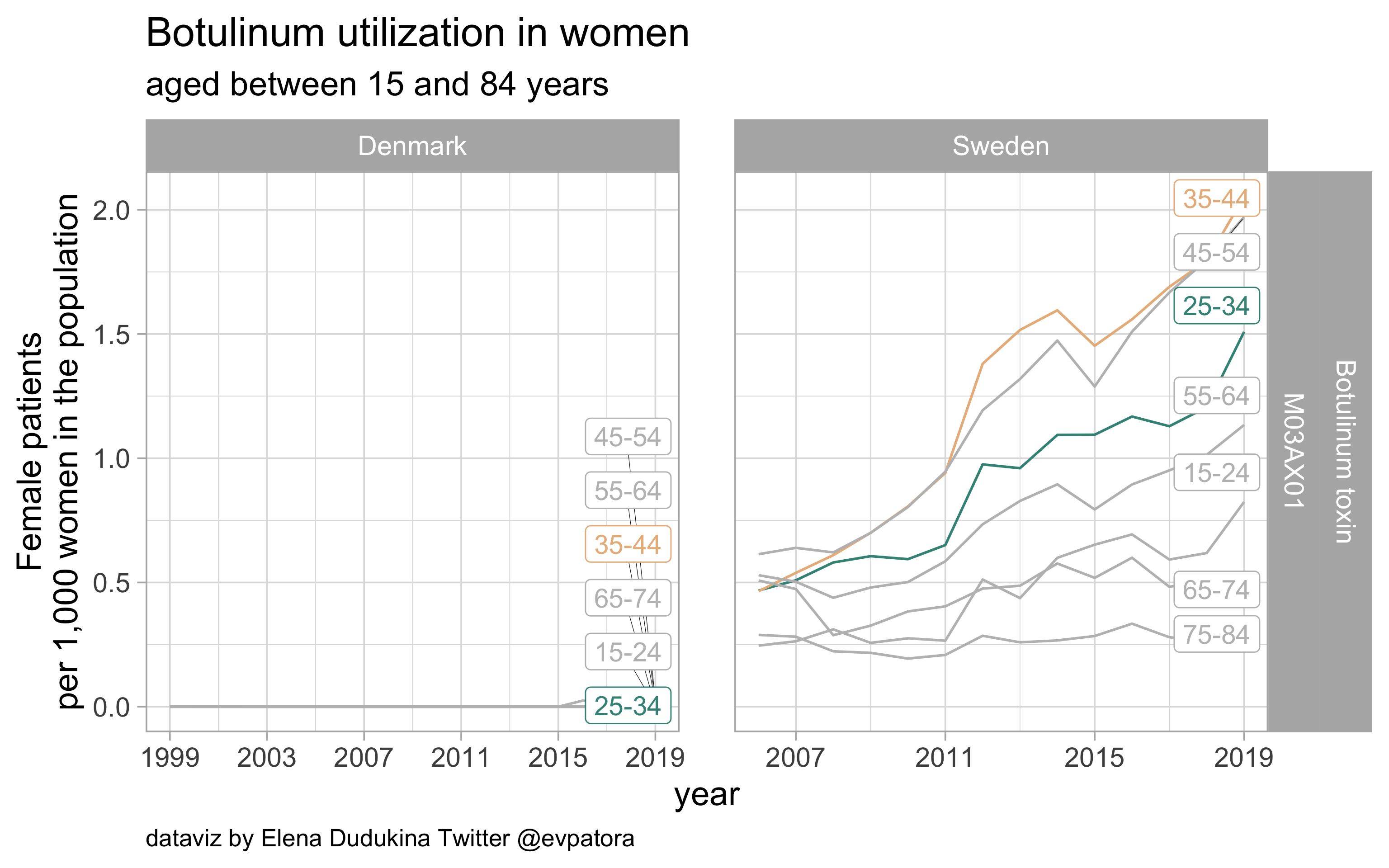
results_all %>%
filter(str_detect(ATC, regex_botulinum)) %>%
kableExtra::kable(format = "html")
| ATC |
drug |
age_cat |
Denmark |
Sweden |
| M03AX01 |
Botulinum toxin |
25-34 |
0.000000 |
1.508491 |
| M03AX01 |
Botulinum toxin |
35-44 |
0.017775 |
2.052792 |
Conclusion
In the publicly available aggregated prescription data, the utilization of different medications with a possible indication for treating migraine episodes among women 25-34 and 35-44 years was mostly comparable between Denmark and Sweden. However, utilization patterns of some drugs (eg, triptans, non-steroids overall and specifically naproxen, paracetamol, salycilic acid and caffeine, amitriptyline) noticeably differed between the two Nordic countries.
Cheers! ✌️




















